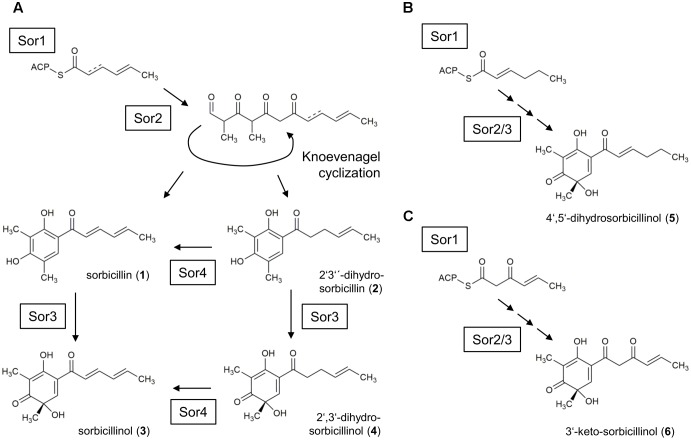
FIGURE 9.
Model for the sorbicillinol biosynthesis pathway in T. reesei. (A) The basic model is identical to the earlier proposed model by Fahad et al., 2014 (compare Figure 1). Briefly, the PKS Sor1 iteratively combines three acetyl units and the growing chain is modified by the ketoacyl reductase subunit, and optional by the enoyl reductase subunit in the second cycle. The polyketide is handed over to the PKS Sor2, which adds three more acetyl units, and two methyl groups. Sor2 releases an aldehyde, which undergoes spontaneous cyclization resulting in the formation of sorbicillin (1) or 2′,3′-dihydrosorbicillin (2). Next, the FAD-dependent monooxygenase Sor3 transforms sorbicillin or 2′,3′-dihydrosorbicillin to sorbicillinol (3) or 2′,3′-dihydrosorbicillinol (4). Further, Sor4 might reduce 2′,3′-dihydrosorbicillin (2) to sorbicillin (1) and/or 2′,3′-dihydrosorbicillinol (4) to sorbicillinol (3). The obtained data from our study (Figures 4–6) suggest that the main product of Sor1/2 is in fact 2′,3′-dihydrosorbicillin. (B) Assumed that the enoyl reductase subunit of Sor1 was active during the first cycle, 4′,5′-dihydrosorbicillinol (5) would have been synthesized by Sor2 and Sor3. (C) Assumed that the ketoacyl reductase subunit of Sor1 was not active during the third cycle, Sor2 and Sor3 would have assembled 3′-keto-sorbicillinol (6).