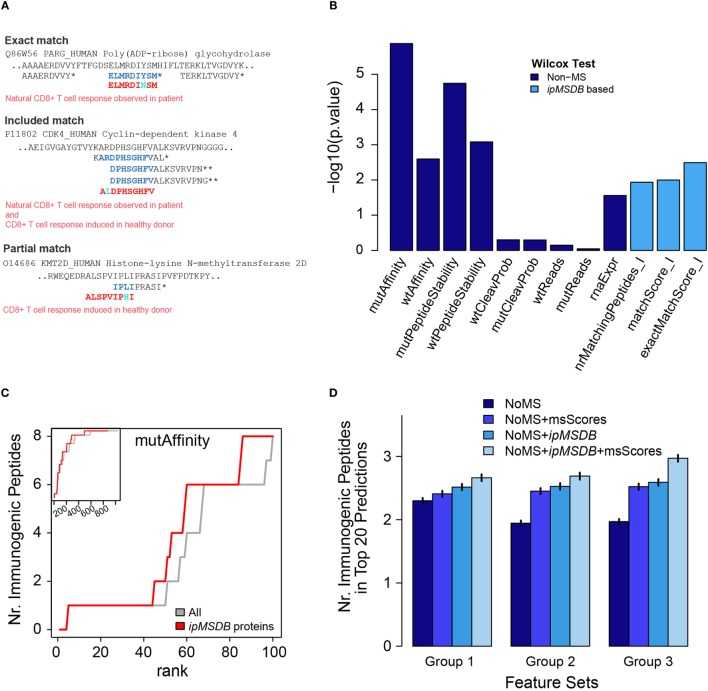
Figure 5.
(A) Examples of matches of confirmed immunogenic neoantigens from Stronen et al., with peptides in ipMSDB. Predicted neoantigen sequences are in red, overlapping amino acids within peptides detected by mass spectrometry (MS) are in blue and mutation are in cyan. Matches can be “exact,” signifying that the exact wild-type (wt) counterpart of the neoantigen sequence was detected by MS, “included,” i.e., the neoantigen is included within the sequence of a longer wt peptide detected by MS, or “partial,” i.e., the neoantigen is partially overlapping at the position of the mutation with the wt counterpart detected by MS. *HLA-I peptides, **HLA-II peptides. (B) The higher the −log10 of Wilcox-test p-values, (i.e., the smaller the pValue), the more different the distribution of immunogenic peptide values is to the distribution of non-immunogenic peptide values. See Ref. (41) for details about the non-MD scores. (C) Predicted peptides were ranked by mutAffinity score (HLA-binding affinity of mutated peptide) and the number of correctly predicted immunogenic peptides (maximally 16) is plotted against the rank. Gray line represents before and red line after removal of proteins not present in ipMSDB. The main figure shows the first 100 ranks, whereas the inset shows all 1,034 ranks with the y-axis ranging from 0 to 16. (D) Average number of correctly predicted immunogenic peptides in the top 20 peptides ranked by support vector machine regression. Vertical small bars represent ± 2 times the standard deviation of the average values. msScores are exactMatchScore_I and matchScore_I. In group 1, non-MS scores were mutAffinity, mutPeptideStability, and rnaExpr. In group 2, the non-MS scores were the scores from group 1 plus wtAffinity and wtPeptideStability that take into consideration also aspects related to the wt peptide counterparts. In group 3, non-MS scores were the scores from group 1 plus diffAffinity, diffPeptideStability, which incorporate the differences in the binding affinity and in the binding stability, respectively, of the wt and the mutant peptides. diffAffinity = mutAffinity−wtAffinity and diffPeptideStability = mutPeptideStability−wtPeptideStability.