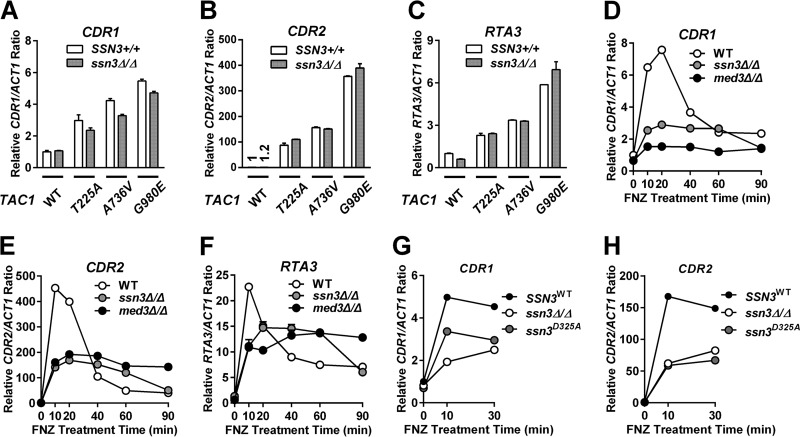
FIG 10.
Expression of TAC1 target genes in ssn3 deletion and kinase-dead mutant strains. (A to C) RT-qPCR analysis of CDR1 (A), CDR2 (B), or RTA3 (C) expression in SSN3+/+ strains with individual TAC1 variants (wild type [DSY2937-35], T225A [ACY67], A736V [ACY13], or G980E [ACY71]) and their ssn3Δ/Δ derivatives (yLM236, yLM237, yLM238, and yLM239, respectively). The expression level of each gene measured in the SSN3+/+ strain with wild-type TAC1 (DSY2937-35) was set to 1. The CDR2 expression levels in DSY2937-35 and yLM236 are denoted above each corresponding column. (D to F) RT-qPCR analysis of fluphenazine-induced expression of CDR1 (D), CDR2 (E), and RTA3 (F) in an ssn3Δ/Δ strain with wild-type TAC1 (yLM236). Parallel analyses of fluphenazine induction in a wild-type (WT; DSY2937-35) and a med3Δ/Δ mutant (yLM232) strain are presented for comparison. The basal expression level of each gene in DSY2937-35 was individually set to 1. (G and H) RT-qPCR analysis of fluphenazine-induced CDR1 (G) and CDR2 (H) expression in an ssn3Δ/Δ null strain (yLM265) complemented by either wild-type SSN3WT (yLM279) or a kinase-dead allele SSN3D325A (yLM276). The CDR1 and CDR2 expression levels in the SSN3WT strain (yLM279) collected before treatment were individually set to 1.