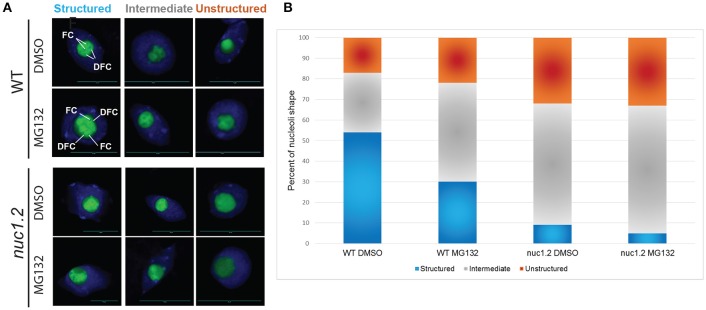
Figure 4.
MG132 proteasome inhibitor affects nucleolus structure. (A) Nucleoli from WT and nuc1.2 plants, expressing the Fib2:YFP constructs, treated or not with 50 μM MG132. 100 nucleoli were analyzed in each sample. Green fluorescence of the Fib2:YFP, is used to visualize nucleolus organization with the FC (Fibrillar Centers), and the DFC (Dense Fibrillary Component) components in Structured, Intermediate and Unstructured forms. DAPI staining was used to visualize nucleoplasm and distinguish the nucleolus. (B) The bar graph depicts the percentage of structured (blue), intermediate (gray) and unstructured (orange) nucleoli in WT and nuc1.2 plants treated or not with 50 μg MG132. Reactions without proteasome inhibitors (DMSO only) were used as control. 100 nuclei for each plants and conditions were analyzed. Scale bar = 10 μm.