Figure 1. Overall structure of the KATP channel bound to ATP and GBC.
(A) Linear sequence diagram for the Kir6.2 and SUR1 polypeptides, with primary domains colored to match the panels below. Numbers indicate residue number at the beginning and end of each domain. (B) Cryo-EM density map of the KATP channel complex at 3.63 Å resolution, viewed from the side. Gray bars indicate approximate position of the bilayer. (C) View of map from extracellular side. (D). Structural model of the complex, with ligands ATP (green) and GBC (red) in boxes. (E) View of the model from the extracellular side.
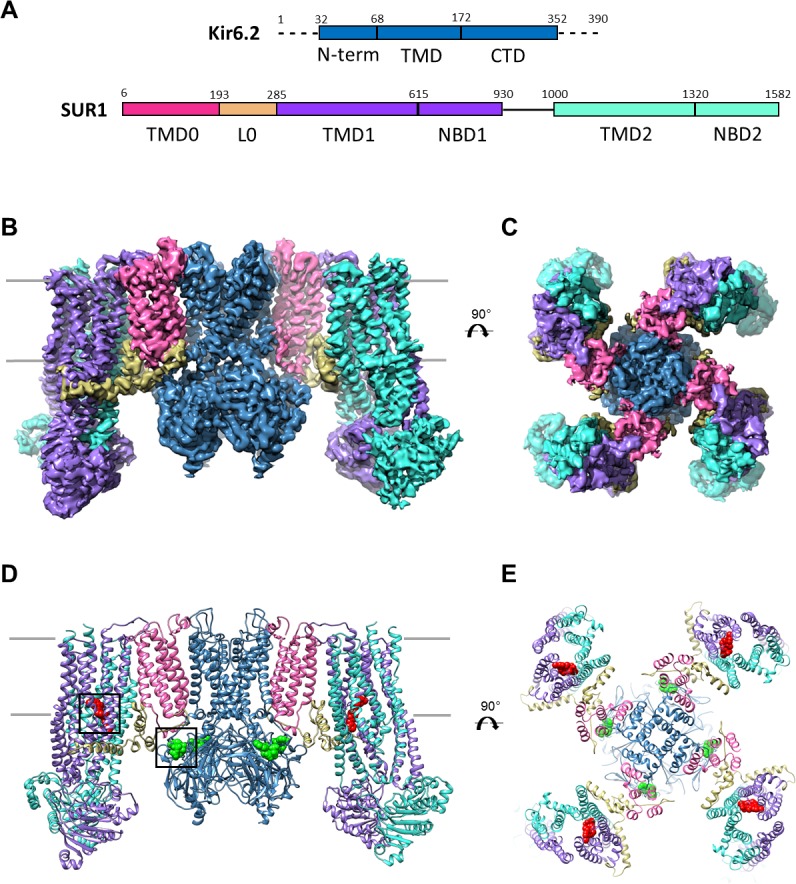
Figure 1—figure supplement 1. Data collection and image processing workflow.
Figure 1—figure supplement 2. Cryo-EM density map analysis.