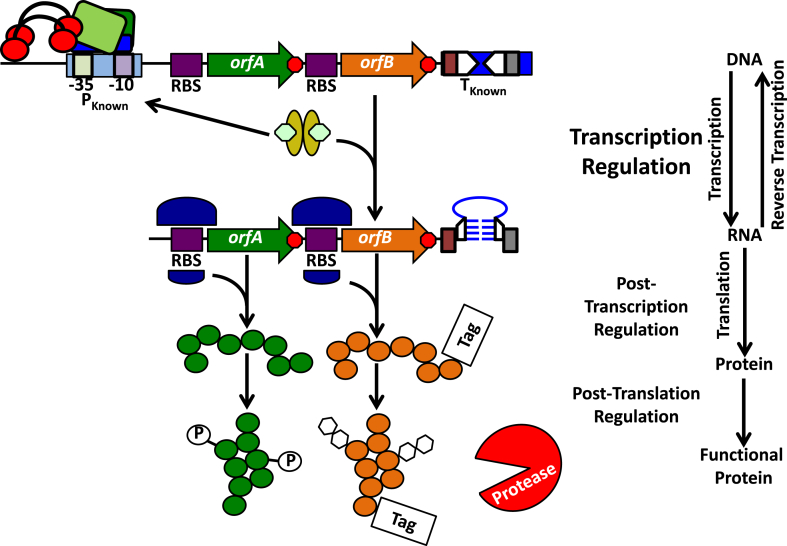
Fig. 1.
Overview of the central dogma of molecular biology. An operon encoding orfA and orfB is under the control of the indicated promoter (PKnown), which is engaged by the RNA polymerase holoenzyme (green and blue boxes, red circles, and black arcs). Sequences that will be transcribed into the ribosome binding sites (RBSs) for orfA and orfB (purple boxes) and the transcription terminator (TKnown; two inverted repeats [inverted arrowheads] flanked by a poly A-tract [red box] and poly U-tract [grey box]) are indicated. Red octagons represent translation stop codons. At the level of transcription regulation, alterations to the promoter and terminator sequences, the presence of transcription activators and repressors (depicted as golden ovals with green hexagon ligands bound), and nucleoid/supercoiling states may all influence orfA and orfB transcription (not shown). Through the process of transcription itself, the associated orfA and orfB mRNA is produced, which may in turn be bound by ribosomes (depicted as two variable size purple half circles) at the RBS. To terminate transcription, the depicted terminator sequence forms a stem-loop (blue), mediated by base pairing/stacking with the complementary sequences of the former inverted repeats (half arrowheads), and mRNA synthesis ceases at this site (poly A- and ploy U-tracts are indicated as above). Modifying RBS strength, RNA riboswitches, aptazymes (aptamer ribozymes), and RNA stability may each contribute to post-transcription regulation (not shown). Folding of a translated protein (green and orange circles) with the assistance of folding chaperones (not shown), enzymes that promote post-translation modifications such as phosphorylation (P in a white circle) or glycosylation (white hexagons), the presence of proteolytic enzymes (indicated in the figure and potentially targeted to a protein through a specific motif [Tag]) represent methods of post-translation regulation.