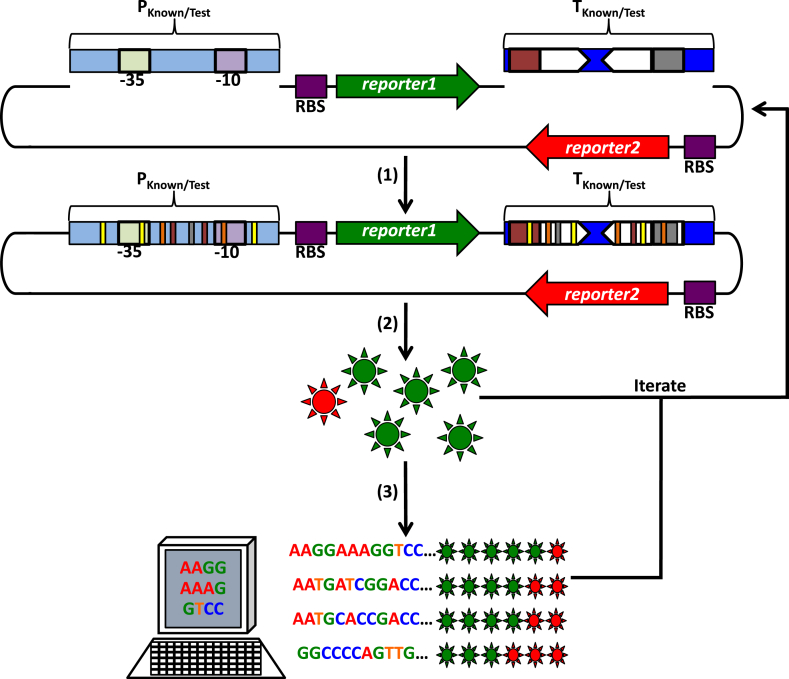
Fig. 3.
A procedure to generate promoter and terminator libraries. (1) To test putative promoters as sites of transcription initiation, known promoters (PKnown) or random sequences (PTest) may be inserted upstream of a previously promoterless ribosome binding site (RBS)-reporter1 gene translational fusion (purple box with a green arrow). These promoters may be constitutive (shown) or subject to transcription regulation by activators and repressors (not shown). Pale green and pale purple boxes indicate the −35 and −10 promoter sequences, respectively. To test putative transcription terminators, known terminators (TKnown) or random sequences (TTest) may be inserted upstream of a promoterless RBS-reporter2 translational fusion (purple box with a red arrow) and downstream of the PKnown/Test-reporter1 transcription fusion above. Additional known or random terminators may be inserted in tandem, and a known terminator may be inserted downstream of reporter2 to ensure termination of transcription after RNAP reads-through this gene (not shown). Inverted white arrows represent the sequence forming a terminator stem-loop (inverted repeats), and red and grey boxes represent poly A- and poly U-tracts, respectively. (2) To introduce variability in promoter and terminator libraries (thin yellow, orange, red, grey, and white bars) these cis-regulatory elements may be derived from randomly sheared DNA, synthesized on demand with desired sequences, subjected to mutagenic PCR (polymerase chain reaction), or other similar methods. These cis-regulatory element constructs may be introduced into an organism of interest, reporter gene expression may be assessed for each library member (green and red suns), and this procedure may be iterated (starting with new random/mutated sequences or using library member sequences as inputs for further sequence variation generation) to achieve a desired result or a wider expression variability profile. (3) To identify sequence characteristics that may be responsible for reporter gene expression level differences (rows of green [gene expression from promoters] and red [gene expression from transcription read-through] suns) among library member sequences (indicated), these constructs should be sequenced and compared with other members or known promoter sequences (iterated as desired).