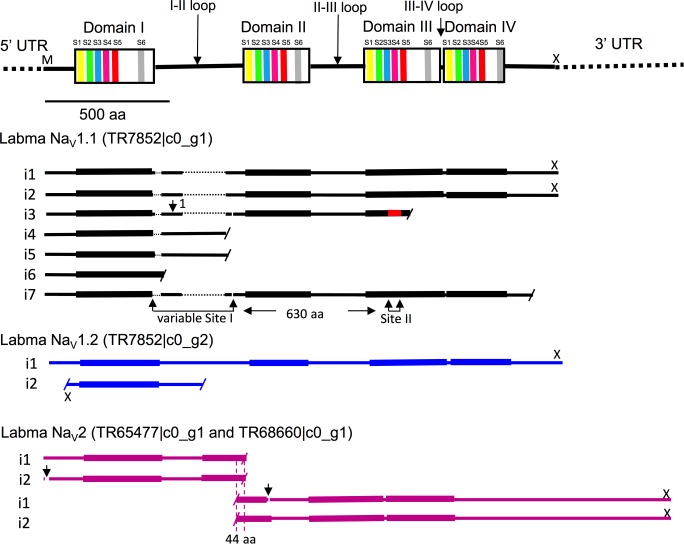
Fig 4. Labidocera madurae voltage-gated sodium channel sequences assembled by Trinity.
Diagram at top shows the four well-conserved domains (DI-DIV) bridged by less-well-conserved loops. Conserved domains are depicted vertically expanded to show approximate locations of six trans-membrane α-helical segments (colored bands labeled S1, S2-S6). Sodium-selectivity of the NaV1 transcripts (but not NaV2) is confirmed by the occurrence of four characteristic amino acids (aspartic acid, glutamic acid, lysine and alanine [DEKA]) in specific locations termed the "P-loops" [31]. Coverage by variants of three putative genes, Labma NaV1.1 Labma NaV1.2 and Labma NaV2 indicated by bars labeled with the i number assigned by Trinity. For Labma NaV1.1, no one sequence possessed all of the pieces (putative exons), so the overall span across the diagram represents a manual reconstruction generated by including all of the pieces from the different i’s. Gaps in sequences are indicated by fine dotted lines. Identical 5' (504 nucleotide) UTRs for i1-i7 have been omitted, as have the identical 3' UTRs (1518 nucleotides) of i1 and i2. Within each gene, corresponding residues across different i’s were identical (reflected in the same coloration of the bars) in almost all cases, except for the splice variant indicated in red for NaV1.1 i3. Sequences representing partial predicted proteins not initiated by an M at the N-terminal or terminated by a stop codon (“X” above the bar) at the C-terminal are indicated with a short diagonal bar. Positions of the domains for NaV2 differ somewhat from those of NaV1 shown in the top diagram and are indicated by thickening of the bars. Two sites of putative splice variation (Site I and II) are indicated below the NaV1.1 diagram, and one non-optional segment within Site I is designated "1" (96aa). Arrows in the NaV2 diagram indicate short optional pieces (gaps in the horizontal bars), and the overlap region between the two pairs of isoforms of 44 identical amino acids (aa) is indicated.