Figure 2.
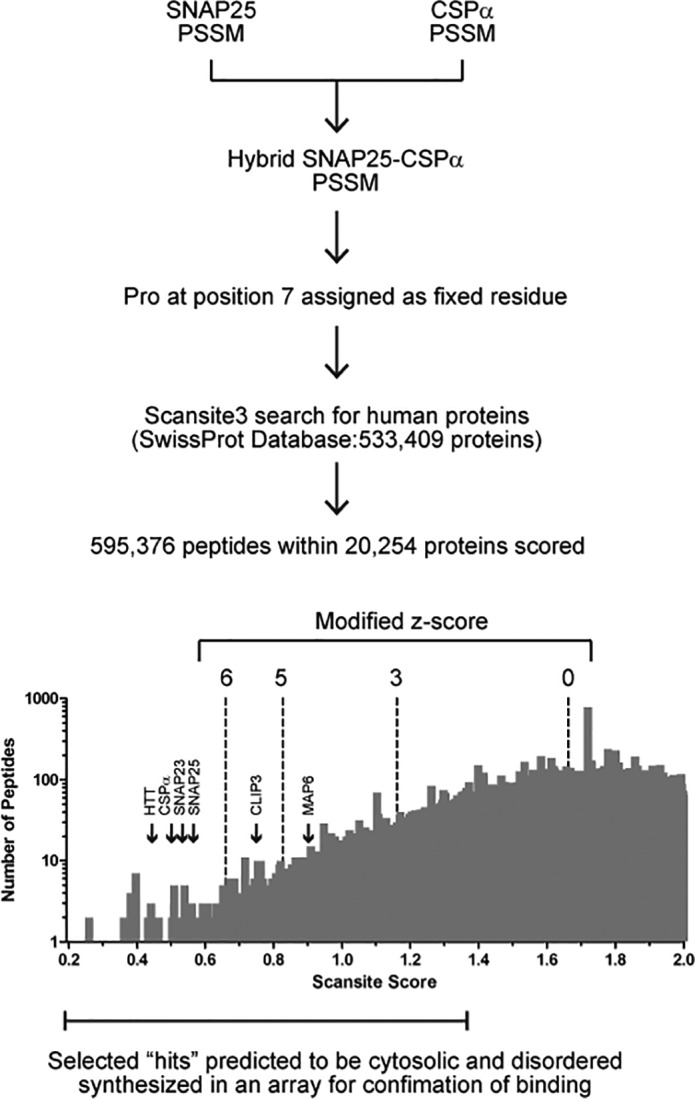
Procedure used for the in silico prediction of zDHHC-AR-binding motif (zDABM) sequences across the human proteome. A hybrid SNAP25 and CSPα PSSM was created, from averaging scores from individual SNAP25 and CSPα PSSMs (derived from quantification of far-Western blots shown in Fig. 1) and adjusting this PSSM for Scansite, with Pro at position 7 assigned as a fixed amino acid (see ”Experimental procedures“ for details). The derived PSSM was used for matching peptides across the human proteome (SwissProt database; UniProt release 2011_11). The number of peptides scored and the distribution of peptides for each score are shown. Selected hits with Scansite scores ranging between 0.2 and 1.4 (see ”Experimental procedures“ for more details) were filtered for cytosolic localization and disorder prediction to be included for validation of ARzD17 binding. Peptides with a modified z-score (number of S.D. below the median Scansite score −1.663) above 6 were considered as high confidence hits. The positions in the histogram of the six peptides that have been previously shown to bind to the AR of zDHHC17 and zDHHC13 are shown.
