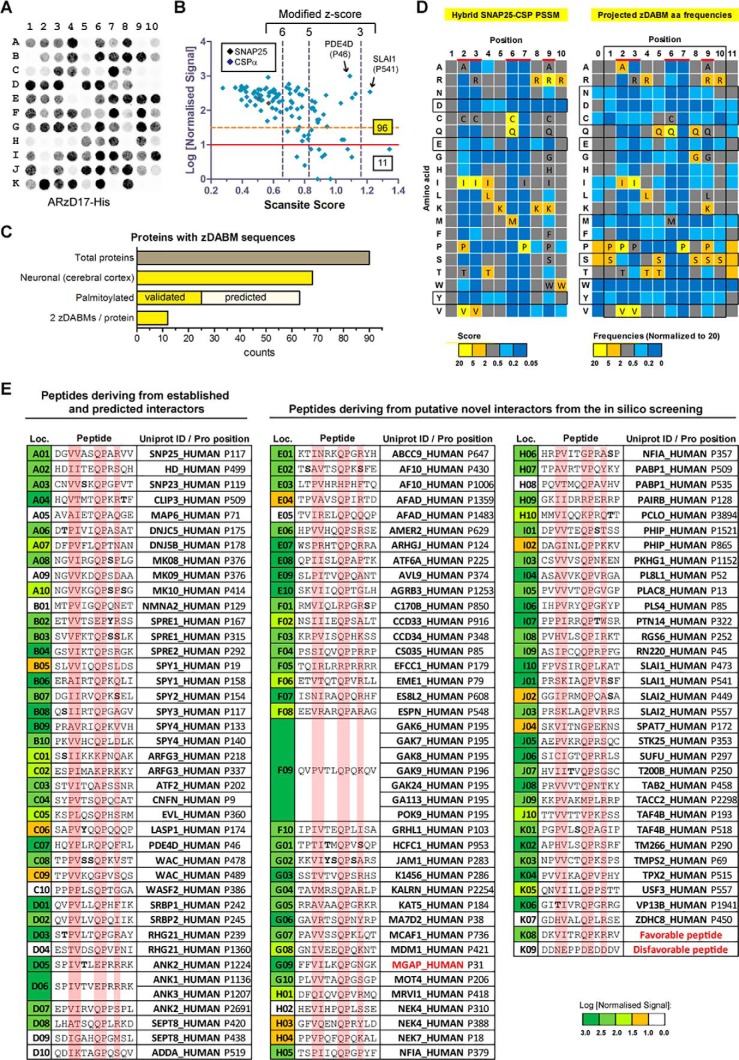
Figure 3.
Validation of predicted zDABM sequences. A peptide array was constructed comprising a total of 109 12-mer peptides, which correspond to 107 human sequences, and two artificial sequences: one expected to bind ARzD17 (favorable peptide) and one not expected to bind (disfavorable peptide). Binding to ARzD17 was assessed by far-Western blotting using purified human ARzD17-His (51–288 amino acids) and detection with a histidine tag antibody. Quantified signals were normalized to the highest observed signal, which was given an arbitrary value of 1000; peptides with values less than 10 were considered non-binders. A, peptide array far-Western blot. B, normalized signal intensities were plotted against the Scansite-derived score. Peptides below the red line (log values <1) were considered as non-binders; whereas peptides below the dashed orange line (log values <1.5) were considered as weak binders. The total numbers of binding and non-binding peptides, as well as the position of SNAP25 and CSPα in the plot, are shown. C, bar graph showing the total number of proteins containing zDABM sequences and the number that are known to be neuronal, palmitoylated (SwissPalm database), or predicted to be palmitoylated (palmitoylation sites were predicted by CSS Palm 3.0 using high threshold setting), as well as the number of proteins that contain two zDABM-binding sites as opposed to one site (the nuclear protein MGAP was excluded from these analyses). D, comparison between the amino acid preferences defined by the hybrid SNAP25-CSPα PSSM, and the amino acid preferences defined by the projected amino acid frequency for each position of zDABM-binders. For the projected amino acid frequencies, the remaining high-confidence sequences were also considered on top of all the natural peptides identified as zDABM sequences. The more critical binding positions are highlighted in red, with all but disfavorable amino acids for these positions shown; favorable-only amino acids for all other positions are indicated too. Amino acids within the motif that appear highly disfavorable (in at least 7/10 positions) or favorable (in at least 5/10 positions) are highlighted in a box. E, peptides used at the corresponding locations on the peptide array, with the most critical for binding positions highlighted; corresponding peptide positions for each peptide within human proteins are shown. Residues that are known to be modified by phosphorylation (PhosphoSite database) are indicated in bold. Non-natural zDHHC17-interactors (proteins that are only known to be nuclear and artificial peptide sequences) are shown in red.