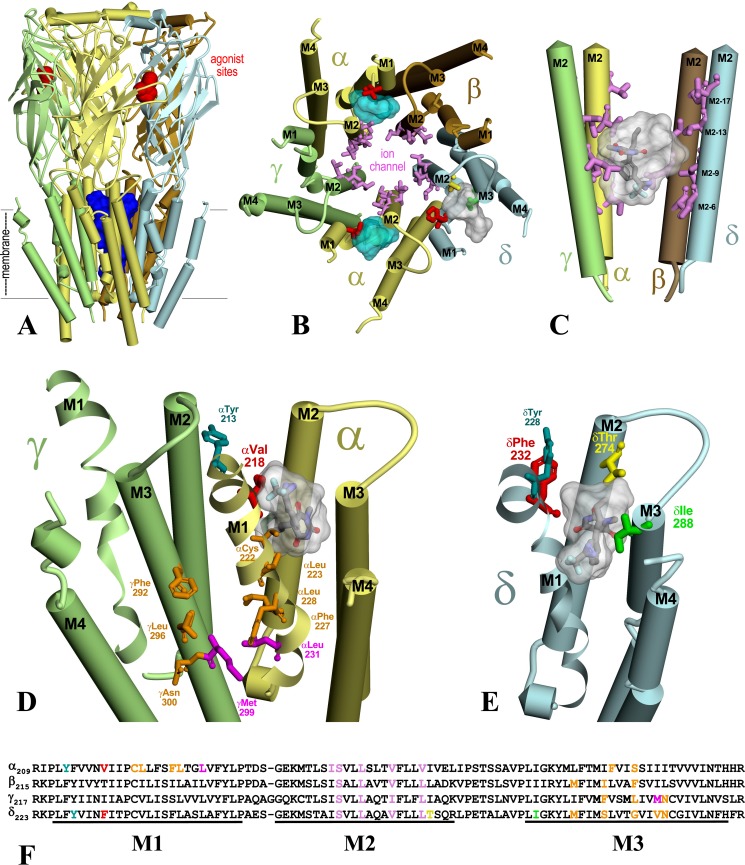
Figure 10.
S-mTFD-MPPB binding sites in the Torpedo nAChR. A T. californica nAChR homology model was constructed based on the crystal structure of human (α4)2(β2)3 nAChR (Protein Data Bank entry 5KXI (16)). A, side view of the nAChR extracellular and transmembrane domains (α (yellow), β (brown), γ (green), and δ (light blue)) with nicotine (red Connolly surface) in the ACh-binding sites and the ion channel in blue. B, a view of the nAChR TMD from the base of the extracellular domain. C, the binding site in the ion channel. D and E, views from the lipid of the γ–α subunit interface (D) and the δ subunit TMD (E), at a tilt angle optimizing visualization of the α and δ subunit helix bundle pockets. The amino acids photolabeled by [3H]S-mTFD-MPPB are shown in stick representation in the ion channel (B and C; pink), in the α subunit helix bundle (B and D; αVal-218 (red)), and in the δ subunit helix bundle (B and E; δPhe-232 (red), δThr-274 (yellow), and δIle-288 (green)). In C–E, the locations of S-mTFD-MPPB (molecular volume = 269 Å3) docked in the binding sites are shown in stick representations (carbon (gray), hydrogen (white), oxygen (red), nitrogen (blue), and fluorine (cyan)) in the most favorable binding mode and/or as Connolly surface representations of the volumes defined by the ensemble of the 10 most energetically favorable binding poses. Also highlighted in D and E are the amino acids photolabeled by [3H]R-mTFD-MPAB (magenta, αLeu-231 and γMet-299 (26)) at the γ–α interface, by [14C]halothane (teal, αTyr-213 and δTyr-228 (43)) in the helix bundle pockets, and by [125I]TID (orange, αCys-222, αLeu-223, αPhe-227, αLeu-228, γPhe-292, γLeu-296, and γAsn-300) at the lipid interface. F, subunit sequence alignment for the M1–M3 region, with the same color coding of amino acids as shown in B–E to identify photolabeled residues.