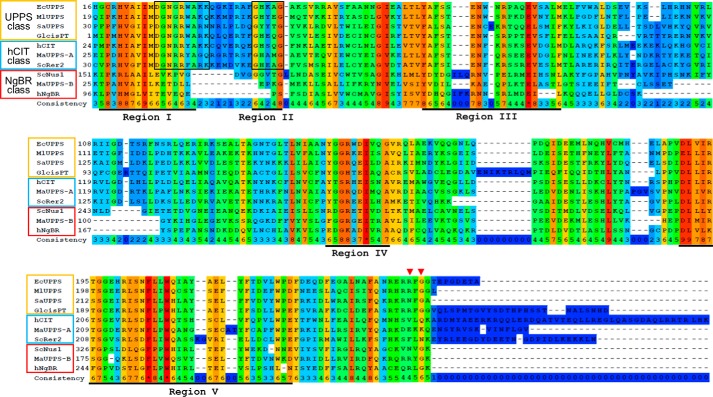
Figure 1.
Multiple alignment of the proteins bearing cis-PT homology domain. Proteins represented in the figure are single subunit cis-PTs: EcUPPS (E. coli, GenBankTM accession no. P60472), MlUPPS (M. luteus; GenBankTM accession no. BAA31993.1), SaUPPS (Sulfolobus acidocaldarius; GenBankTM accession no. WP_011277635.1), and Glcis-PT (G. lamblia; GenBankTM accession no. XP_001709868.1); orthologs of hCIT cis-PT subunit: hCIT (human, GenBankTM accession no. BAB14439), ScRer2 (S. cerevisiae, GenBankTM accession no. P35196), and MaUPPS-A (M. acetovorans, GenBankTM accession no. AAM07075.1); and orthologs of NgBR cis-PT subunit: NgBR (human, GenBankTM accession no. NP_612468), ScNus1 (S. cerevisiae, GenBankTM accession no. NP_010088), and MaUPPS-B (M. acetovorans, GenBankTM accession no. WP_011024281.1). The conservation scoring is performed by PRALINE. The scoring scheme works from 0 for the least conserved alignment position, up to 10 (*) for the most conserved alignment position. Positions of five conserved regions originally described among cis-PTs are underlined. Residues involved in catalysis and substrate binding conserved between hCIT orthologs and single subunit cis-PTs are boxed. Red triangles indicate the positions of conserved arginine/asparagine and glycine in the C-terminal RXG motif of NgBR orthologs and homomeric cis-PTs.