Figure 1.
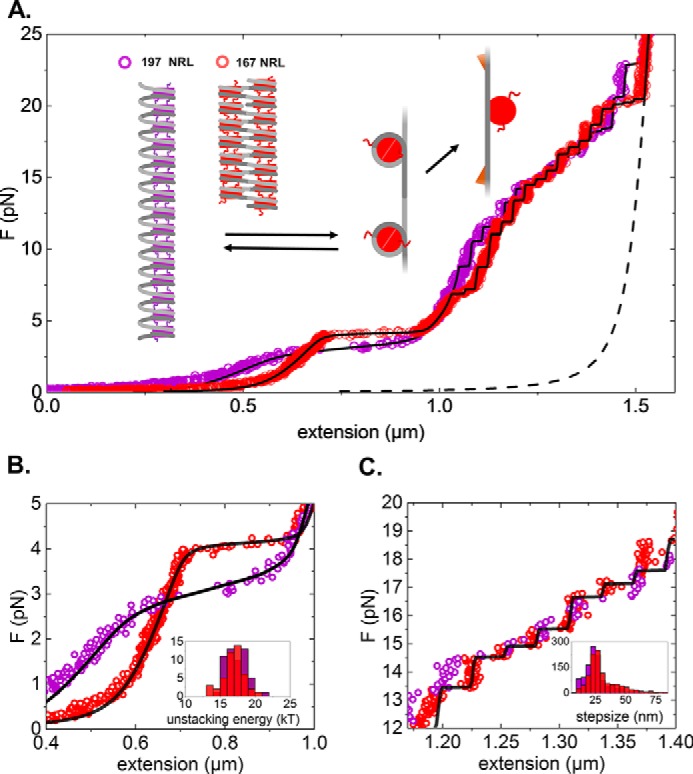
Wild-type histone chromatin fibers with 167- and 197-bp NRLs respond differently to applied force. A, force (F)-extension curve of a chromatin fiber reconstituted on 15 × 167-bp DNA (20-bp linker DNA; red) and 15 × 197-bp DNA (50-bp linker DNA; violet). As a reference, the WLC extension of a 4.4-kbp DNA is plotted (black dotted line). Fitted black curves correspond to the statistical mechanics model for chromatin unfolding described in Meng et al. (33). The graphs represent conformational changes upon stretching. A folded fiber initially stretches linearly until nucleosome-nucleosome interactions rupture and simultaneous release of 56 bp of nucleosomal DNA. The remaining nucleosomal DNA releases in two steps, a small 5-nm step around 6 pN and an ∼25-nm step at higher forces. Note that tetrasomes only feature the last step. B, zoom of the low-force part of the force-extension curve, representing the stretching of the folded fiber. Compared with 197-bp NRL fibers, 167-bp NRL fibers are more condensed (although extra tetrasomes or hexasomes in this fiber shift the curve to higher condensation), are stiffer, have a higher rupture force, and rupture cooperatively. The unstacking transition is in equilibrium, and the fiber refolds as the force is released (data not shown). Inset, histogram of the fitted unstacking energy per nucleosome for both types of fibers. C, zoom of the high-force regime of the force-extension diagram. The remaining wrapped DNA unfolds in a non-equilibrium fashion, resulting in a staircase-like extension curve. Inset, histogram of the step sizes.
