Figure 7.
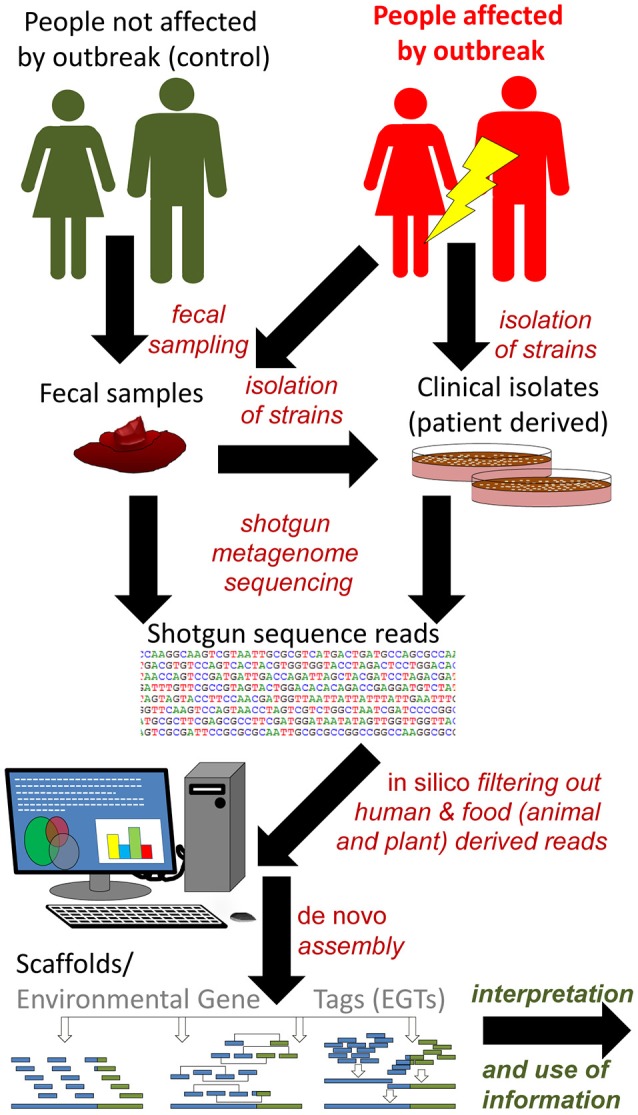
Reference independent shotgun metagenome sequencing based approach for identification and characterization of outbreak strain(s). In case of smaller outbreaks the possibility to compare metagenomes from affected people (patients) and healthy controls is limited. In these cases the availability of clinical isolates may be required to avoid exhaustive open ended bioinformatics (in silico) analyses, as exemplified by Brzuszkiewicz et al. (2011) and Rasko et al. (2011). Environmental gene tags (EGTs) from metagenomes of people affected by the outbreak and controls (people not affected) can be compared in case of a larger outbreak, as exemplified by Loman et al. (2013). In that study, EGTs present only in affected patients were characteristic of the outbreak strain and provided sufficient information to near complete characterization of its genome. Scaffolds and in particular assembled genomes may and should be uploaded to reference database(s), for successive use in analytical approaches like those described in Figure 6.
