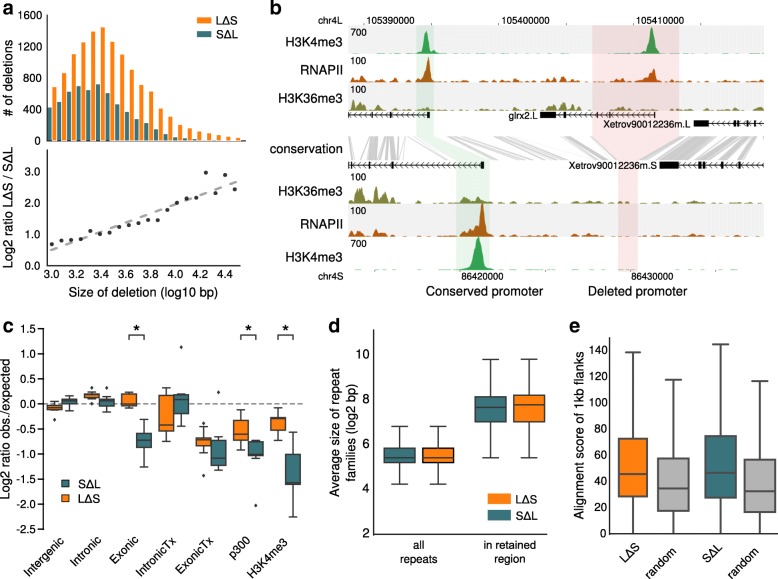
Fig. 3.
The S subgenome has more and larger deletions than L. a Size frequency distribution of deletions (top) and size ratio of LΔS deletions relative to SΔL deletions as a function of deletion size (bottom). b An example of a gene (grlx2) that has lost the promoter on the S genome due to a deletion. Shown are the gene annotation (black), ChIP-seq profiles for H3K4me3 (green), RNAPII (brown), and H3K36me3 (dark green). The sequence conservation is indicated by gray lines. c The log2 fold difference between the observed number of deleted basepairs and the expected number (mean of 1000 randomizations). The fold difference is calculated per chromosome and summarized in a boxplot. Intergenic 1 kb distance from a gene, Intronic introns, Exonic UTRs + CDS, IntronicTx introns from genes actively transcribed, ExonicTx exons from genes actively transcribed, p300 genomic fragments having a p300 peak, H3K4me3 genomic fragments having a H3K4me3 peak. The asterisks mark significant differences between the L and S chromosomes (p < 0.001, Mann–Whitney U test). d Retained regions associated with deletions are enriched for relatively long repeats (p < 1e-52 for both LΔS and SΔL; Mann–Whitney U test). e 1 kb flanks of the retained regions are more similar to each other than random genomic regions of the same size (p < 1e-114 and 1e-83 for LΔS and SΔL, respectively; Mann–Whitney U test)