Fig. 1.
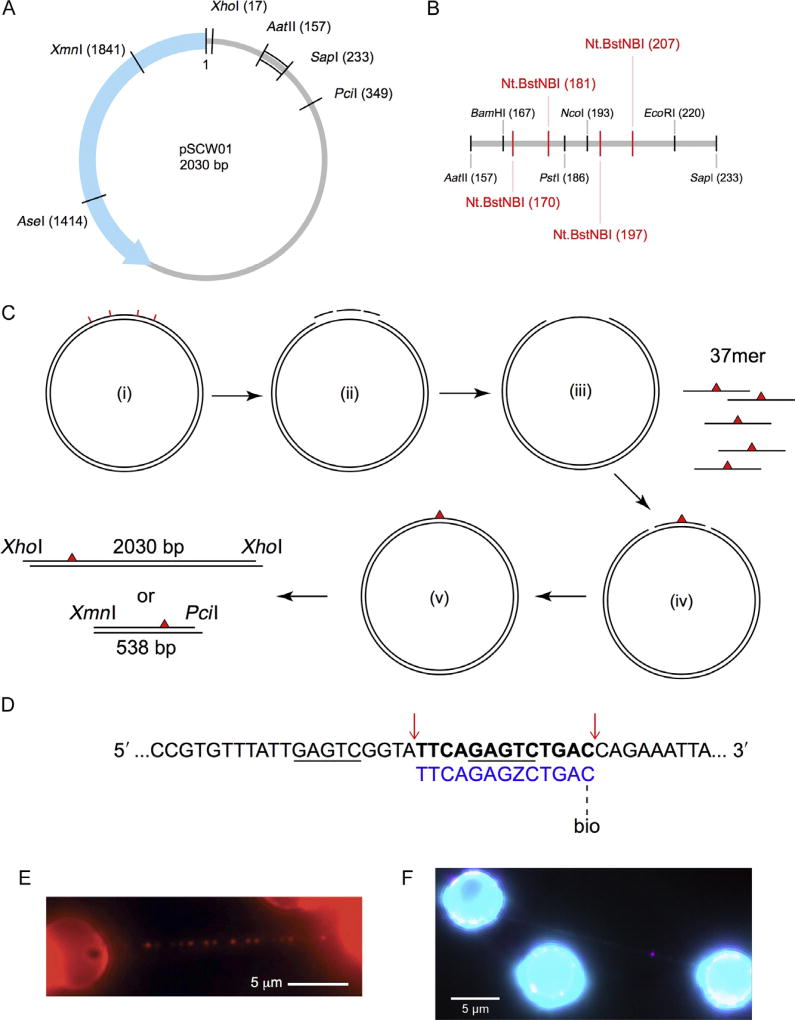
Design of defined lesion substrate. (A) Map of the pSCW01 plasmid and locations of restriction digest sites. (B) Detailed map of restriction digest and nicking sites for plasmid sequence between the AatII (157) and SapI (233) sites. Nt.BstNBI nicking sites are shown in red. (C) Strategy for generating defined lesion substrate based on the pSCW01 plasmid. The plasmid is first nicked by Nt.BstNBI at four different locations (i), which yields three short single-stranded fragments (ii) that are liberated from the plasmid via heating, resulting in a gapped plasmid (iii). 37mer oligonucleotides, each containing a site-specific lesion, are annealed to the gapped plasmids (iv) before the nicks on either side of the oligonucleotides are sealed by overnight ligation (v). The plasmids can now be linearized by XhoI and then tandem-ligated to form long DNA substrates, containing one site-specific damage per 2030 bp, for use in the DNA tightrope assay. Alternatively, the plasmids can be double digested by XmnI and PciI and gel purified to obtain 538 bp fragments, each containing one site-specific damage ~160 bp from the PciI site. (D) Strategy for inserting a damaged oligonucleotide with a biotin conjugate for quantum dot visualization in λ-DNA. The upper λ-DNA sequence is underlined at the binding sites for Nt.BstNBI. Cut sites are indicated by red arrows, leading to the release of the bolded segment. This is replaced by the lower 5′ phosphorylated oligonucleotide (blue) containing damage (Z = fluorescein-dT) and biotin-conjugated via TEG at the 3′ end. (E) An array of streptavidin-conjugated quantum dots on a DNA tightrope of a defined lesion substrate containing one site-specific abasic site analog per 2030 bp, each with a proximal biotin marking the site of the lesion. (F) DNA damage (magenta) visualized with 655 streptavidin-conjugated quantum dot on a λ-DNA tightrope stained with YOYO-1 (cyan). Panel E: Adapted with permission from Ghodke, H., Wang, H., Hsieh, C. L., Woldemeskel, S., Watkins, S. C., Rapic-Otrin, V., et al. (2014). Single-molecule analysis reveals human UV-damaged DNA-binding protein (UV-DDB) dimerizes on DNA via multiple kinetic intermediates. Proceedings of the National Academy of Sciences of the United States of America, 111(18), E1862–E1871. http://dx.doi.org/10.1073/pnas.1323856111 (fig. 4A).