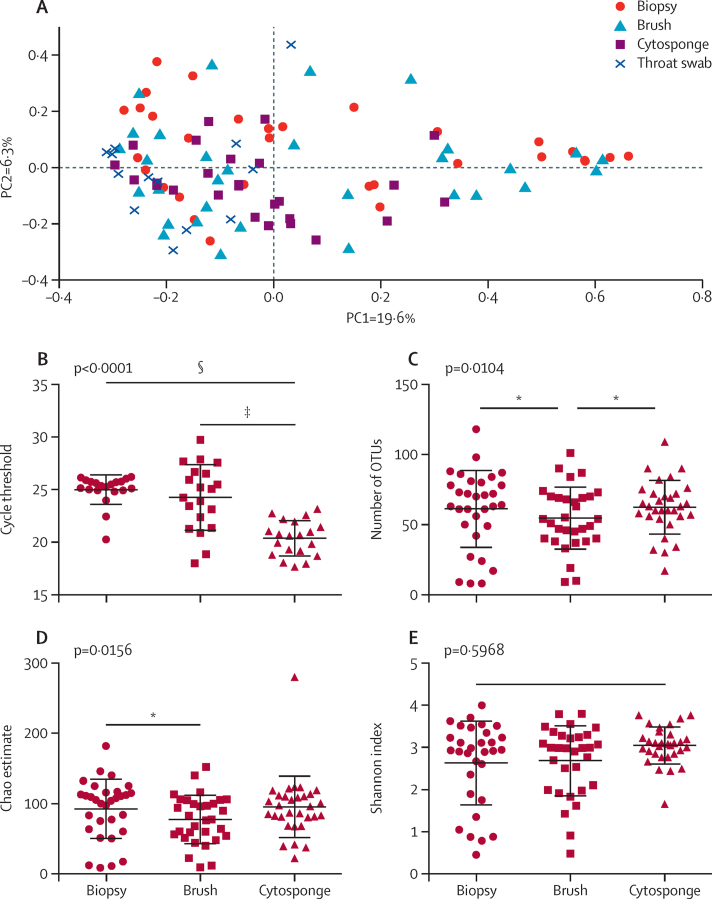
Figure 4.
Comparison of different methods to sample the oesophageal microbiota
(A) Principal coordinate analysis with the Bray-Curtis algorithm for matched endoscopic biopsies, brushes, and Cytosponge samples (31 patients) and 13 throat swabs from a subset of these patients. The first axis (PC1) accounts for 19·6% of the sample variance and the second axis (PC2) accounts for 6·3% of the variance. Data were subsampled at 631 reads per sample. (B) Overall bacterial abundance using 16S rRNA gene-based quantitative PCR in matched endoscopic biopsies, brushes, and Cytosponge samples (20 patients), Friedman test and Dunns multiple comparisons post test. (C) The observed diversity of bacterial operational taxonomic units (OTUs), (D) the Chao estimate of total OTU richness, and (E) the Shannon diversity index for matched endoscopic biopsies, brushes, and Cytosponge samples (31 patients), Friedman test and Dunns multiple comparisons post test. Data were subsampled at 631 reads per sample. *p<0·05. †p<0·01. ‡p<0·001. §p<0·0001.