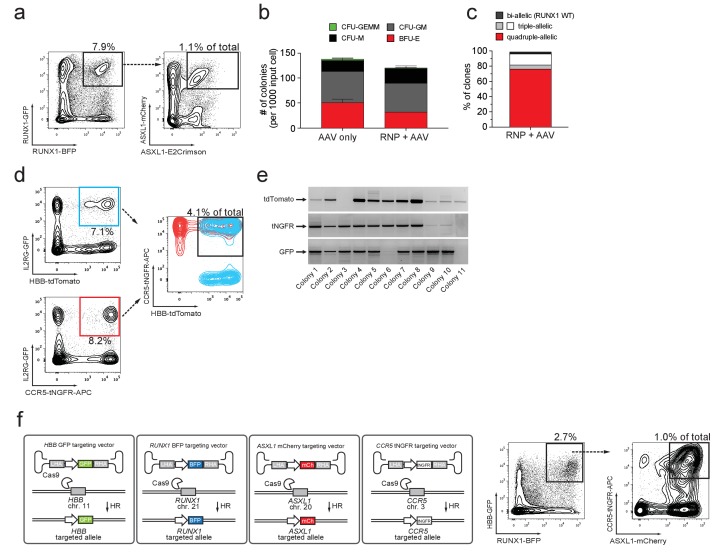
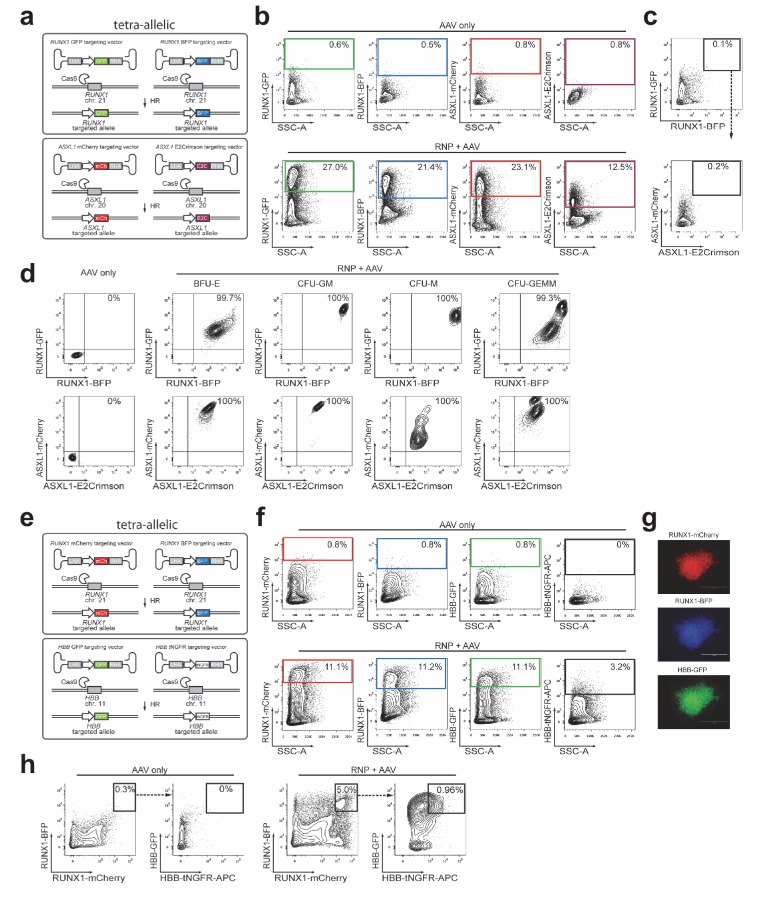
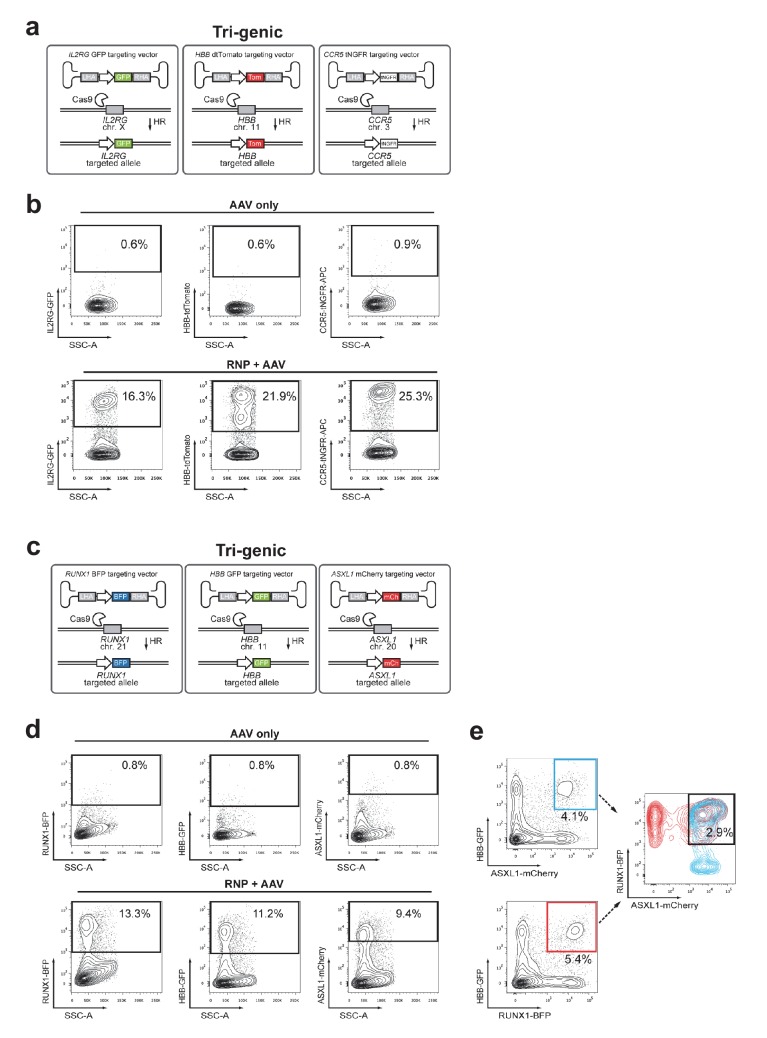
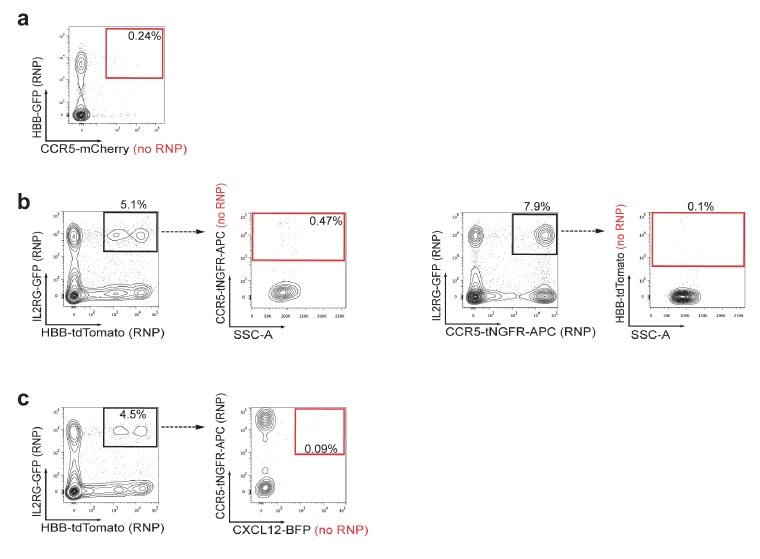
Figure 4. Multiplexing homologous recombination in CD34+ human hematopoietic stem and progenitor cells (HSPCs).
(a) HSPCs were electroporated with Cas9 RNP targeting ASXL1 and RUNX1 followed by rAAV6 transduction with two donors for ASXL1 (mCherry and GFP) and two donors for RUNX1 (E2Crimson and BFP). Tetra-allelically targeted HSPCs were identified as mCherryhigh/GFPhigh/BFPhigh/E2Crimsonhigh (N = 3 see Supplementary file 1e) (b) Cells modified at both alleles for RUNX1 and ASXL1 (as in (a)) were subjected to a methylcellulose assay (triplicates) and scored as BFU-E, CFU-M, CFU-GM or CFU-GEMM based on morphology 14 days after sorting. (c) PCR was performed on colony-derived gDNA to detect targeted integrations at both genes. 73 individual colonies were analyzed. Color coding for colonies with triple-allelic integration are as follows: grey: RUNX1 biallelic/ASXL monoallelic; white: RUNX1 monoallelic/ASXL1 biallelic. (d) For tri-genic targeting of HSPCs, cells were electroporated with Cas9 RNP targeting IL2RG, HBB, and CCR5 followed by transduction of three rAAV6 donors homologous to each of the three genes (IL2RG-GFP, HBB-tdTomato, and CCR5-tNGFR). Tri-genic-targeted cells were identified as reporterhigh for all three reporters (N = 5 see Supplementary file 1e). (e) Methylcellulose clones from the triple-positive cells in (d) were subjected to genotyping PCR and gel images show colonies with targeted integration at all three genes in 9/11 colonies (note that GFP shows a faint band in colony 6). (f) Left, Schematic showing strategy for targeting four different genes (HBB, RUNX1, ASXL1, and CCR5) simultaneously (tetra-genic). Four different genes are targeted by electroporation of four different Cas9 RNPs followed by transduction with four different rAAV6 donors that each targets a gene with a different reporter. Right, Tetra-genic targeting at the above-mentioned four genes was identified as reporterhigh for all four reporters (N = 3 see Supplementary file 1e).