Fig. 3.
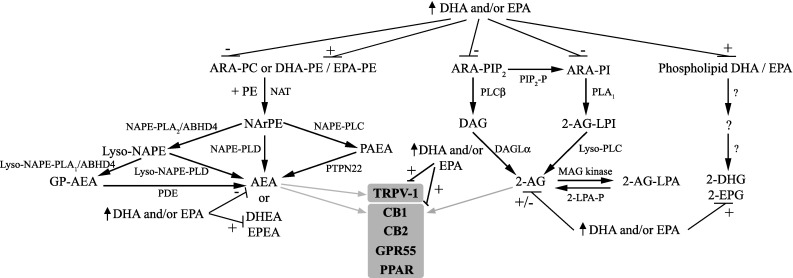
Interplay in the synthesis and actions of the 2-acylglycerols and ethanolamides derived from ARA, DHA and EPA. The major pathway for AEA production begins with N-acyltransferase (NAT) transferring ARA from phosphatidylcholine (ARA-PC) to phosphatidylethanolamine (PE) to generate N-arachidonoyl phosphatidylethanolamine (NArPE), which is followed by hydrolysis by N-acyl phosphatidylethanolamine-selective phospholipase D (NAPE-PLD) to produce AEA. Further pathways include NAPE deacylation by the α/β-hydrolase domain containing 4 (ABHD4) and either the glycerophosphoarachidonoylethanolamide produced (GP-NAPE) cleaved by phosphodiesterase (PDE) to produce AEA or lyso-NAPE is hydrolysed by lyso-NAPE-phospholipase D (PLD) directly to AEA. NAPE can also be hydrolysed by phospholipase C (NAPE-PLC) to generate phospho-anandamide (PAEA), which is dephosphorylated to AEA by phosphatases such as protein tyrosine phosphatase (PTPN22). DHEA and EPEA production from phospholipid bound DHA and EPA appears to share the same pathways. Synthesis of 2-AG occurs from phosphatidylinositol-bound ARA (ARA-PI) via phospholipase C-β (PLCβ) and production of an ARA-diacylglycerol (DAG), which is hydrolysed by diacylglycerol lipases-α to produce 2-AG. Further pathways include dephosphorylation of 2-AG-lysophosphatidic acid (2-AG-LPA) by LPA phosphatase (2-LPA-P) or via phospholipase A1 (PLA1) converting PI to 2-arachidonoyl-lyso PI (2-AG-LPI) and then to 2-AG by lyso phospholipase C (lyso-PLC). The pathways of 2-DPG and 2-EPG production are currently unknown. 2-AG and AEA act at CB1 and CB2 receptors, GPR55 and PPAR, with AEA additionally acting at TRPV-1 (shown in grey). Dietary DHA and EPA enrichment decreases phospholipid ARA and increases phospholipid DHA and EPA, and favours production of DHA and EPA-derived endocannabinoids, whereas acute DHA and EPA treatment in vitro increases 2-AG. DHA and EPA also regulate CB1, CB2 TRPV-1 and PPAR receptor activity and levels. For detailed explanations, refer to the text
