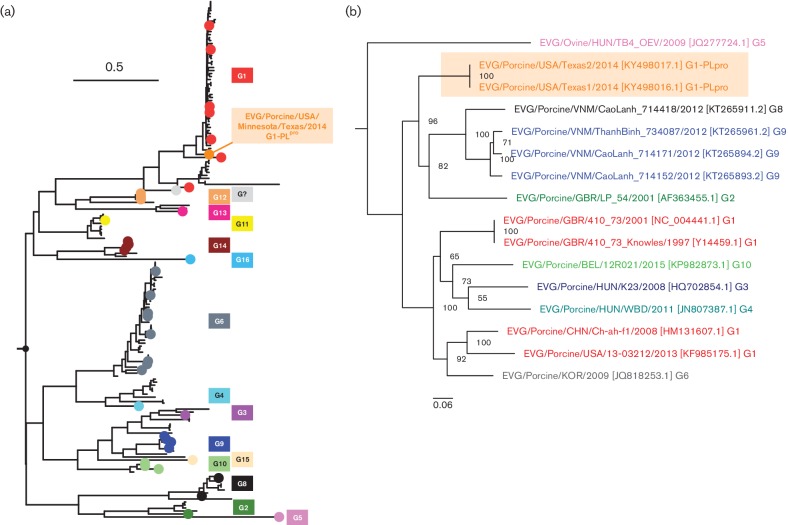
Fig. 2.
Phylogenetic analysis of all currently available EV-G VP1 sequences and complete genomes. (a) Phylogenetic analysis of all currently available VP1 EV-G nucleotide sequences. The tree was rooted at the midpoint and the coloured dots represent taxa that were previously classified using the genotype/serotype nomenclature (www.picornaviridae.com). The branch lengths between taxa are based on the mean number of nucleotide substitutions per site. The VP1 sequences from both Texas strains (orange) fall within the G1 (red) genotype clade. (b) Phylogenetic analysis of full-length EV-G nucleotide sequences. The tree was rooted at the midpoint and the genomes are coloured according to previously published genotypes (i.e. genotypes based on VP1 sequence trees). The strain names are followed by GenBank accession numbers and genotypes. The branch lengths between taxa are based on the mean number of nucleotide substitutions per site and bipartitions are labelled with bootstrapping support values (%). The two Texas strains are highlighted in orange.