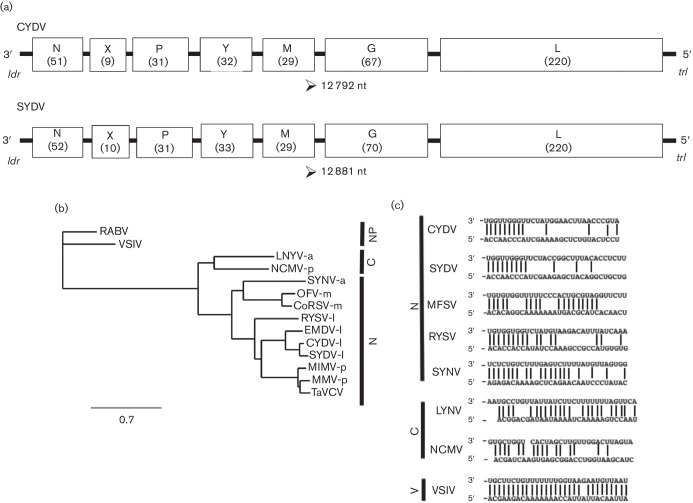
Fig. 1.
(a) Comparison of the CYDV and SYDV genomes. The 12 792 nt CYDV genome is organized into seven ORFs (open boxes) that are separated by conserved gene junctions and flanked by short leader (ldr) and trailer (trl) sequences, respectively. The predicted size of the encoded protein (in kDa) for each ORF is provided in parentheses. (b) A phylogeny of representative negative-strand RNA viruses was inferred from l-protein amino acid sequences. Viruses infecting a variety of hosts were selected, including those that do not infect plants (NP) as well as plant-adapted species that replicate in nuclei (N) or cytoplasm (C) of infected cells. Vectors for the plant-adapted viruses are provided at the end of the virus abbreviation, namely: a, aphid; l, leafhopper; m, mite; or p, planthopper. Virus names and GenBank accession numbers are listed in Methods. CoRSV, coffee ringspot virus; MMV, Maize mosaic virus; TaVCV, Taro vein chlorosis virus; MIMV, Maize Iranian mosaic virus; OFV, Orchid fleck virus; SYDV, Potato yellow dwarf virus-Sanguinolenta strain; CYDV, Potato yellow dwarf virus-Constricta strain; RYSV, Rice yellow stunt virus; SYNV, Sonchus yellow net virus; NCMV, Northern cereal mosaic virus; LNYV, Lettuce necrotic yellows virus; RABV, Rabies virus; VSIV, Vesicular stomatitis virus – Indiana serotype. All branch points had bootstrap values greater than 0.6. The scale bar indicates the number of aa changes per site. (c) Nucleotide complementarity in the leader (3′) and trailer (5′) regions of selected rhabdoviruses in the Nucleorhabdovirus (N), Cytorhabdovirus (C) or Vesiculovirus (V) genera.