Figure 1.
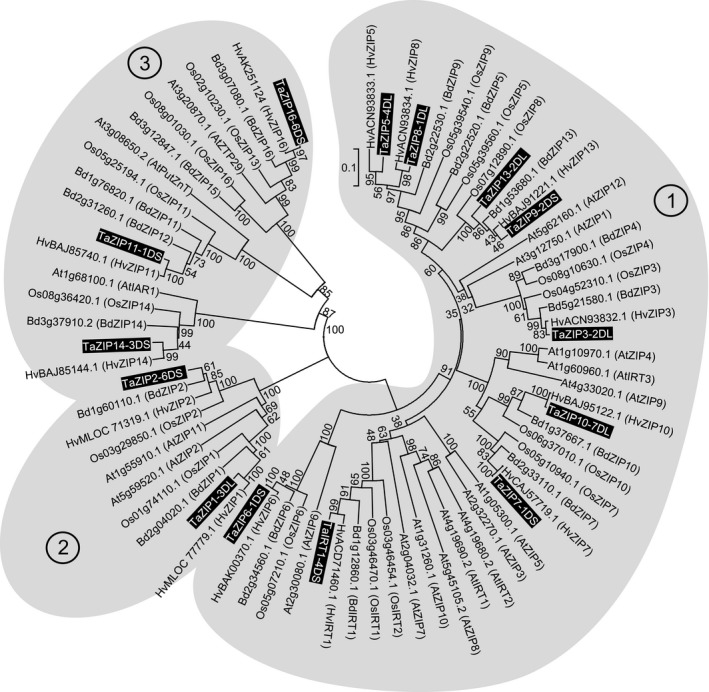
Phylogenetic tree of ZIP proteins from cereals and Arabidopsis.
A neighbour‐joining tree was generated for Arabidopsis (At), rice, Oryza sativa (Os), Brachypodium distachyon (Bd), barley, Hordeum vulgare (Hv), and wheat, Triticum aestivum (Ta) ZIP coding sequence translations. The Muscle algorithm (Edgar, 2004) was used for the alignment of sequences and the phylogenetic tree was created using mega (v.5.2) software. Evolutionary distances were computed using the p‐distance method and are in the units of the number of amino acid differences per site. A thousand bootstrap replicates were used and bootstrap values are shown as percentages. Gene names of Arabidopsis, rice, Brachypodium and barley are shown in brackets, these gene names are in accordance with Tiong et al. (2015). D genome homeologs for T. aestivum are given in this figure. Full accession information for the wheat genes is provided in Table S2.
