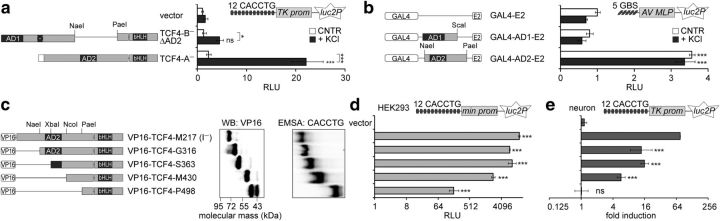
Figure 3.
Mapping of the TCF4 regions required for its regulation by neuronal activity. a, b, d, e, Reporter assays with primary neurons (a, b, e) or HEK293 cells (d) transfected with CMV promoter containing constructs encoding TCF4 isoforms (a), E2-tagged GAL4 fusion proteins (b), or VP16-TCF4-I− deletants (d, e) as indicated. Firefly luciferase constructs carrying μE5 E-boxes (a, d, e) or GAL4-binding sites (b) and Renilla luciferase construct with EF1α promoter were cotransfected. Neurons were left untreated or treated with KCl for 8 h. Data are presented as fold induced levels above the signals obtained from empty vector transfected untreated cells (a, b, d) or as fold induction of luciferase activity in KCl-treated neurons compared with untreated neurons (e). Shown are the means ± SEM from at least three independent experiments (see Materials and Methods for details). Statistical significance shown with asterisks is relative to the reporter levels measured from untreated or KCl-treated cells of the first data point in each chart or between the bars connected with lines. *p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA followed by Bonferroni post hoc tests for comparisons across treatment and transfections (a, b) or one-way ANOVA followed by Dunnett's post hoc tests for comparisons with vector control (d, e). c, Western blot analysis of VP16-TCF4-I− deletants expressed in HEK-293 cell and electrophoretic mobility shift assay (EMSA) for binding of μE5 E-box by in vitro translated VP16-TCF4-I− deletant proteins. AD, Activation domain; AV MLP, adenovirus major late promoter; GBS, GAL4 binding site; min, minimal promoter; RLU, relative luciferase units; TK, thymidine kinase promoter.