Figure 6.
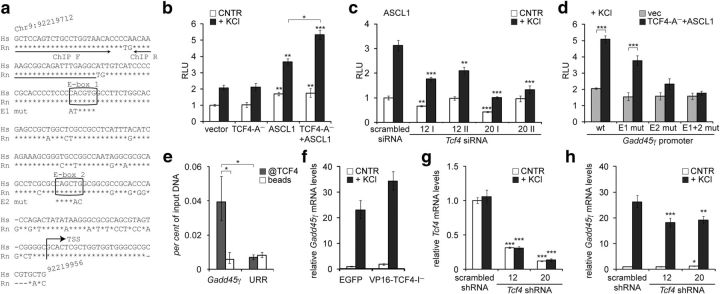
Regulation of GADD45G transcription by TCF4 in neurons. a, Alignment of the cloned GADD45G promoter region in Homo sapiens (Hs) with the respective region in Rattus norvegicus (Rn). Arrows indicate the primers used in ChIP analysis and the putative transcription start site (TSS). The two conserved E-boxes 1 and 2 are shown in boxes with the introduced mutations indicated below. b–d, Reporter assays with cultured primary neurons transfected with WT (b–d) or mutant (d) GADD45G promoter firefly luciferase constructs and Renilla luciferase construct with PGK promoter for normalization. Empty vector, EF1α-TCF4-A− and/or SRα-ASCL1 vector (b, d) and SRα-ASCL1 vector together with TCF4-specific or scrambled siRNAs (c) were cotransfected as indicated. Transfected neurons were left untreated or treated with 25 mm KCl for 8 h. e, ChIP analysis of TCF4 binding to Gadd45g promoter in neurons. Chromatin was coimmunoprecipitated with TCF4 antibodies or with beads alone. The levels of bound Gadd45g promoter or an unrelated region (URR) on chromosome 1 were quantified by qPCR. The means of three independent experiments are presented as percentages of input DNA. f–h, Quantitative RT-PCR analysis of Gadd45g (f, h) and total Tcf4 (g) mRNA expression in primary neurons nucleofected with EGFP or VP16-TCF4-I− constructs (f) or transduced with scrambled or TCF4-specific shRNAs encoding AAV vectors (g, h). Neurons were left untreated or treated with 25 mm KCl for 6 h. b–d, f–h, Data are presented relative to the normalized reporter or mRNA levels measured in untreated control cells in each experiment. The mean results from two (f), three (b, d), four (g, h), or five (c) independent experiments are shown. Error bars indicate SEM. Statistical significance shown with asterisks is relative to the reporter or mRNA levels measured from untreated or KCl-treated neurons of the first data point of each chart or between the bars connected with lines. *p < 0.05; **p < 0.01; ***p < 0.001; two-way ANOVA followed by Bonferroni post hoc tests for comparisons across TCF4 and ASCL1 transfections (b, d) or antibody use and genomic regions (e) or Dunnett's post hoc tests for comparisons with untreated or KCl-treated scrambled si/shRNA samples (c, g, h). RLU, Relative luciferase units.