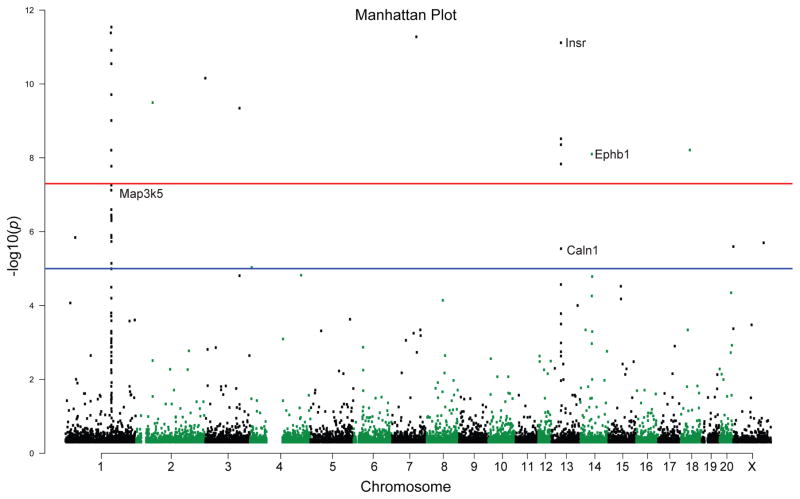
Figure 2.
Manhattan plot illustrating differentially methylated sites in the adult hippocampus of MS-exposed versus control WKY rats. Differentially methylated regions (DMRs) are shown by chromosome (along the x-axis) and the negative logarithm of the association p-value for each block (−log10 [p-value]) on the y-axis). We examined whether DMRs surpassed two thresholds: a) a threshold p-value less than 1 × 10−5, the suggested threshold in genome-wide studies, indicated by a blue line; and then b) a second more stringent threshold p-value less than 5 × 10−8, indicated by a red line. There were 56 DMRs in the MS versus control adult hippocampus that surpassed the lower (blue line) threshold. This group of DMRs included 15 gene regulatory sites (a region within the gene body or up to 10 kb upstream of a gene). When considering the more stringent parameters of the higher (red line) threshold, we found 40 DMRs within 12 gene regulatory sites that passed that threshold. Some example genes that met these criteria are labelled: ephrin type-B receptor 1 (Ephb1), insulin receptor preprotein (Insr), mitogen-activated protein kinase kinase kinase 5 (Map3k5), calcium-binding protein 8 (Caln1)