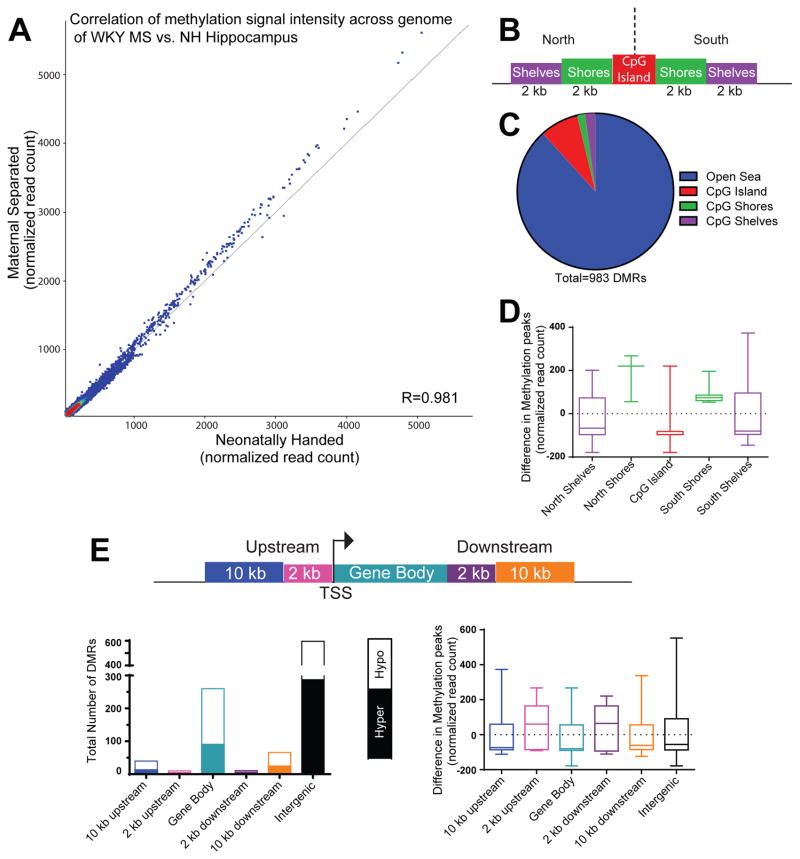
Figure 3.
Early-life maternal separation (MS) exposure elicits lasting DNA methylation changes within several genomic regions of WKY hippocampus. (A) Correlation analysis demonstrated a strong correlation in signal across 308,904 genomic sites that were differentially between samples from MS-exposed and control rats. MS samples represented along the y-axis and neonatal handle control samples along the x-axis. The shift towards increased signal intensity above the diagonal line indicates greater methylation signal in MS versus control hippocampal samples. (B) Graphic depicts an example CpG Island (in red) surrounded by CpG island north and south shores (in green, 0–2 kb from CpG island edges) and CpG island north and south shelves (in purple, 2–4 kb from island edges). Genomic regions >4 kb from any CpG island edge is considered “open sea”. (C) Of the 983 differentially methylated regions (DMRs) between MS and control WKY hippocampal samples, the majority were located in “open sea”, with approximately 12% of the DMRs occurring within CpG islands, shores and shelves. (D) Within the CpG islands and surrounding shores and shelves, sites within the CpG islands, north and south shelves were hypomethylated in MS versus control samples, while sites within north and south shores were hypermethylated between the groups. (E) Methylation data were also classified depending upon where DMRs fell within specific genomic regions, including within: 10 kb or 2 kb upstream of the transcription start site (TSS), the gene body, 2 kb or 10 kb downstream of the gene body. The highest number of DMRs occurred within exons or intergenic regions. On average, sites within regions 2 kb upstream or downstream of a gene body were hypermethylated in MS compared to control samples; DMRs within gene bodies, 10 kb upstream or 10 kb downstream of genes were hypomethylated.