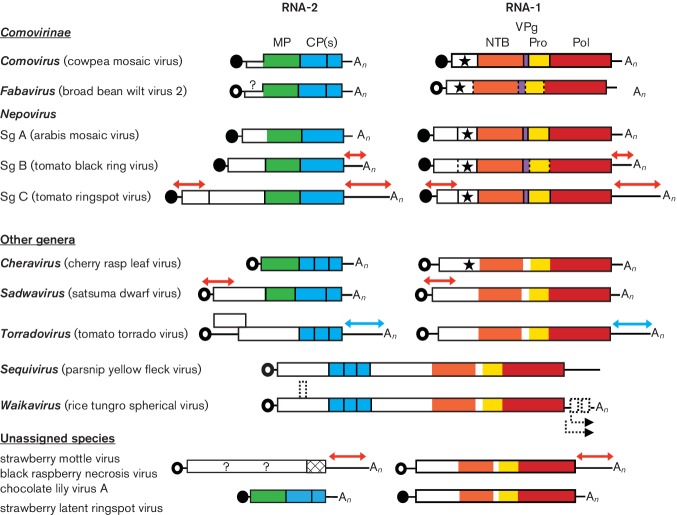
Fig. 2.
Genome organization of representative members of the family Secoviridae. Each RNA is shown with ORFs represented with boxes. Circles depict VPg molecules covalently attached at the 5′-end of the RNAs. Black circles represent VPg confirmed experimentally and open circles represent putative VPgs. Poly(A) tails are represented at the 3′-end of the RNAs when present (An). Red and blue arrows above the sequences represent regions of extensive sequence identity between RNAs 1 and 2. In the latter, for torradoviruses, this identity is also characterized by conserved indels. Protein domains with conserved motifs for the putative NTP-binding protein (NTB, shown in orange), VPg (purple), proteinase (Pro, yellow), RNA-dependent RNA polymerase (Pol, red), movement protein (MP, green) and coat protein(s) (CP, blue) are shown. The star represents a conserved motif found in the protease cofactor (Co-Pro) protein of comoviruses and in the equivalent protein of other viruses. Proteinase cleavage sites identified experimentally or deduced by sequence comparisons are indicated by solid or dotted vertical lines, respectively. Possible ORFs in the genome of waikaviruses are shown with dotted rectangles and putative subgenomic RNAs are shown by dotted arrows below the waikavirus genome. Representatives of each nepovirus subgroup (Sg A, B, C) are also depicted.