Figure 3. Examples of GCR structures deciphered through analysis of NGS data.
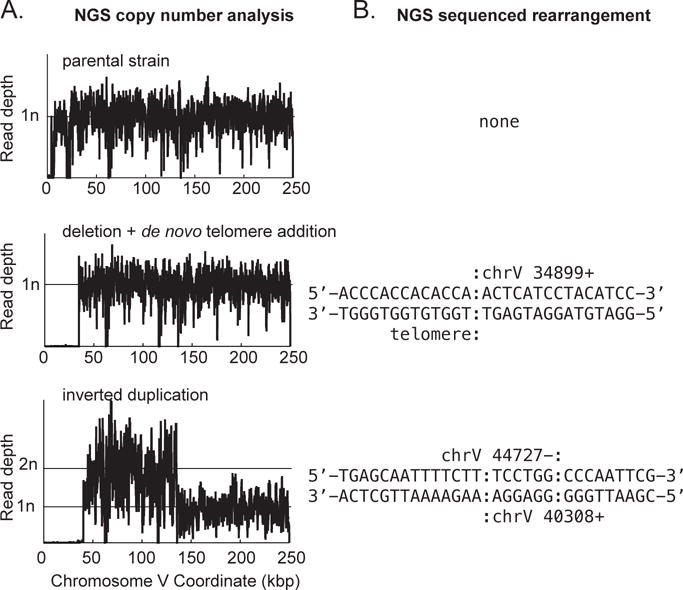
A. Copy number analysis for the left arm of chromosome V is plotted for a parental strain (top), a de novo telomere addition GCR (middle), and a GCR involving an inverted duplication (bottom). Note that both rearranged chromosomes have deleted the region containing the CAN1 URA3 cassette (around coordinate 25,700 in this GCR assay strain). The inverted duplication has additionally duplicated the region from around coordinate 44,700 to the Ty-related sequence YELWdelta6 (around coordinate 138,300). B. Sequence of the novel junctions involved in the observed GCR. A de novo telomere (sequence prior to the colon) healed the terminal deletion at chromosome V 34,899 for the strain displayed in the middle of the panel. For the strain displayed at the bottom of the panel, non-concordant read pairs in the NGS data revealed the presence of an inverted duplication, which was also confirmed by the increase in copy number (panel A) and through the sequence of the reads that are associated with the novel junction and hence did not map to the reference sequence. For this inverted duplication, there is a six base identity (sequence between the colons) at the junction between the source and target of the inversion. The inversion was not a perfect hairpin, however, as ~4,000 bp separate the two fused sequences.
