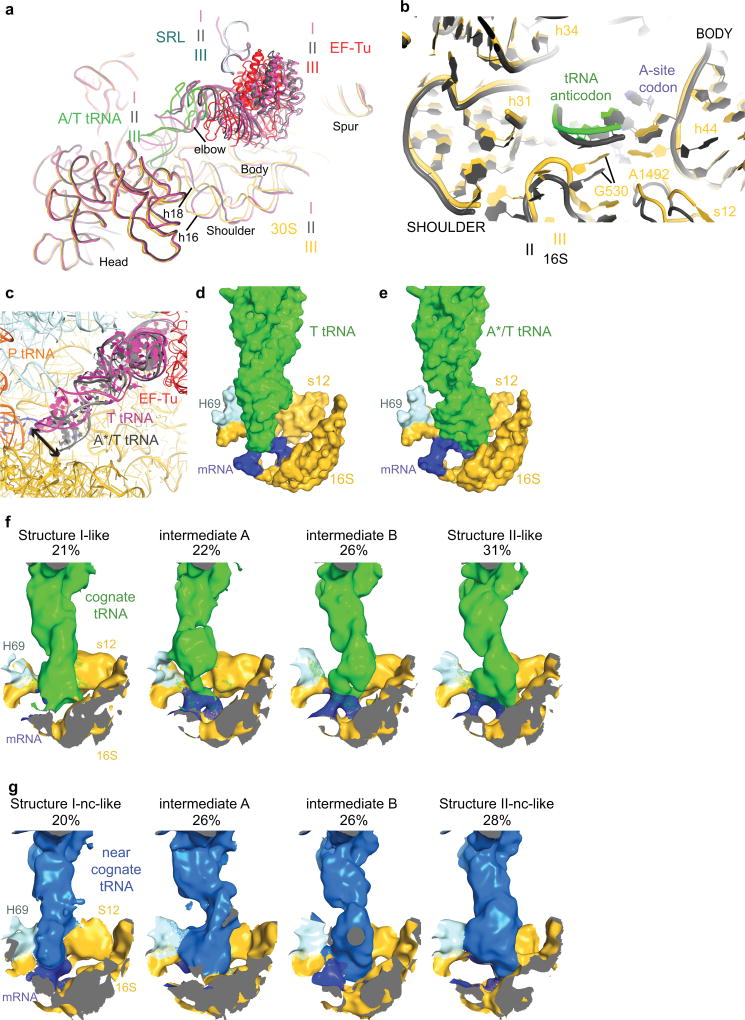
Extended Data Figure 4. 30S domain closure and aa-tRNA conformations in cognate and near-cognate complexes.
(a) Comparison of the 30S conformations among Structures I (magenta), II (gray) and III (multi-colored). Superposition was achieved by structural alignment of 23s rRNA. (b) Superposition of Structure II (gray) and III (multi-colored) highlighting the movement of the shoulder including the 530 loop toward the 30S body including h44. (c) Different conformations of aa-tRNA in Structures I and II: T tRNA (Structure I) is relaxed, whereas A*/T tRNA (Structure II) is kinked to base-pair with mRNA. (d) Interaction of T tRNA in Structure I with the DC is shown in surface representation. All atoms within 15 Å of residues 30–38 of T tRNA are shown except for 16S residues 950–964 and 984–985, which were omitted for clarity. (e) Interaction of A*/T tRNA in Structure II with the DC is shown in surface representation as in (d). (f) Cognate tRNA anticodon samples positions between those in Structure I and Structure II. Additional focused classification into four classes revealed intermediate classes with A-site tRNA density midway between the T tRNA and A*/T tRNA conformations. The cryo-EM density, within 15 Å of residues 30–38 of T or A*/T tRNA, is shown with exceptions as in (d), at 3 σ after applying a B-factor of +200 Å2. (g) Near-cognate tRNA anticodon samples positions between those in Structure I-nc and Structure II-nc. Additional focused classification into four classes revealed intermediate classes with A site tRNA density midway between the T tRNA and A*/T tRNA conformations. The cryo-EM density is shown as in (f).