Figure 2.
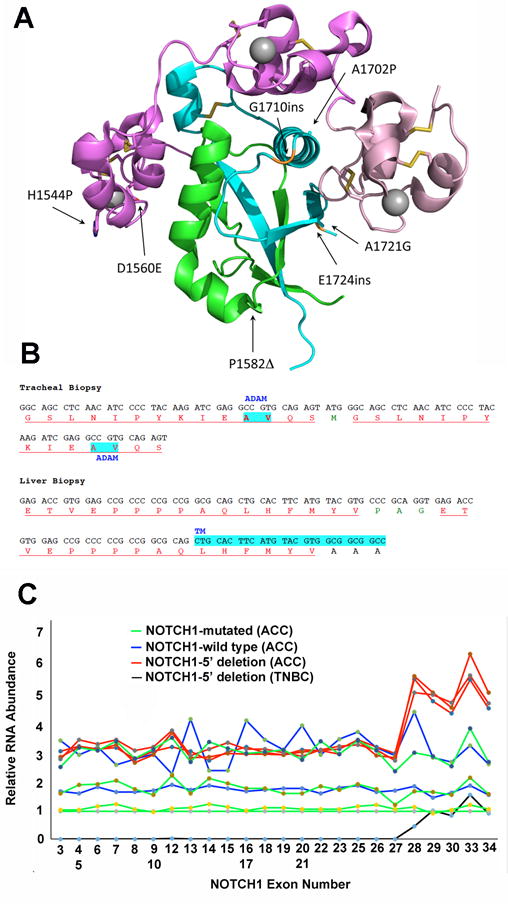
NOTCH1 mutations in ACC. A) Positions of point substitutions and in-frame indels in the NOTCH1 extracellular negative regulatory region (NRR). NOTCH1 structure is modeled based on X-ray crystallographic studies of Gordon et al. (34). B) Sequence of juxtamembrane insertional mutations caused by small NOTCH1 duplications in biopsies of ACC involving the trachea and liver in the same ACC patient. Residues in red correspond to duplicated amino acid residues; residues in green correspond to unique intervening amino acid residues; and residues in black correspond to normal, unduplicated amino acid residues. Highlighted AV residues in the sequence from the tracheal biopsy corresponds to the normal site of ADAM cleavage in NOTCH1, while highlighted residues in the sequence from the liver biopsy corresponds to the NOTCH1 transmembrane domain (TM). C) Nanostring analysis of RNA isolated from FFPE ACC tissue. RNA was isolated from two specimens containing ACCs with wild type NOTCH1 alleles; four ACC specimens with NOTCH1 alleles with point substitutions or small indels (NOTCH1 mutated); three ACC specimens with loss of copy number spanning NOTCH1 exons 1-27; and the triple negative breast cancer cell line MB-157, which has homozygous or hemizygous NOTCH1 rearrangements and lacks NOTCH1 exons 1–27 (13). RNA abundance expressed as Nanostring signal strength (Y-axis) for each exon covered by probes spanning the NOTCH1 locus (X-axis) was first normalized using average signal strength internal control housekeeping genes and then was expressed relative to signal strength for a NOTCH1-mutated ACC with no evidence of allelic imbalance, for which signal strength for each exon was arbitrarily set at 1. Note that excess signals for NOTCH1 exons 28-34, which encode the NOTCH1 transmembrane domain and intracellular domain, is seen in the control MB-157 triple negative breast cancer cell line (black) and in the 3 samples (red) prepared from specimens with copy number loss involving exons 1-27.
