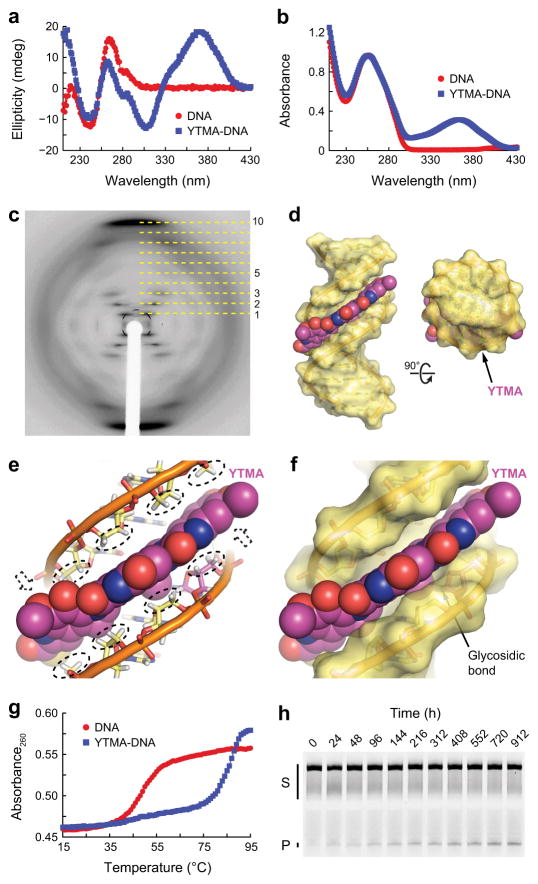
Figure 3. Characterization of YTMA-DNA.
(a,b) Circular dichroism (a) and absorbance (b) spectra of unmodified and yatakemycinylated DNA collected at 15°C. (c) Fiber diffraction pattern from a paracrystalline YTMA-DNA dodecamer. Layer lines are indicated with dashed lines. (d) Hypothetical model of YTMA-DNA. (e) CH/π interactions between YTMA and DNA. Hydrogen atoms that contact the YTM moiety are indicated with dashed ovals or dashed arrows. (f) Close-up of the YTMA-DNA model. (g) Thermal melting profiles of unmodified and yatakemycinylated DNA measured at 260 nm. Melting temperatures [Tm,DNA = 49°C and Tm,YTMA-DNA = 85°C] were calculated by fitting the data to a polynomial function and determining the temperature at which the second-order derivative was equal to zero. (h) Spontaneous depurination of YTMA at 25°C. Full-length YTMA-DNA substrate (S) and cleaved AP-DNA product (P) were separated by denaturing gel electrophoresis (Supplementary Fig. 2). Complete denaturation of the YTMA-DNA duplex was prevented by the high melting temperature of the substrate. Double-stranded YTMA-DNA, single-stranded YTMA-DNA, and YTMA-DNA that denatured during electrophoresis are collectively indicated with a black bar.