Fig 2. Description of eQTLs and identification of candidate variants using CRISPR-Cas9 screening technology.
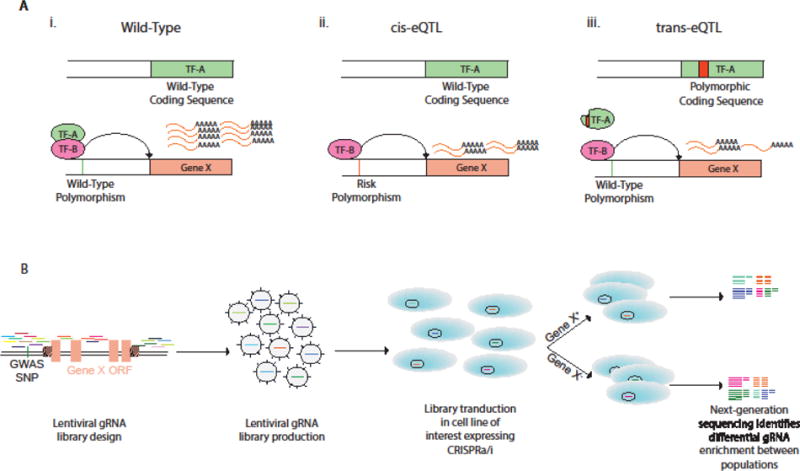
A) Illustration of cis- and trans-expression quantitative trait loci (eQTL). i) Transcription factor A and B (TF-A/TF-B) bind to enhancer elements upstream of Gene X to promote transcription. ii) A polymorphism in the promoter sequence of Gene X inhibits binding of TF-A, reducing transcription of Gene X (cis-eQTL). iii) A polymorphism in the coding region of TF-A prevents TF-A binding to Gene X enhancer element, attenuating expression (trans-eQTL). B) Non-coding CRISPR screen to identify enhancer elements of Gene ×. gRNAs are saturated in non-coding regions flanking and within the open reading frame (ORF). The cell line of interest is transduced with a pool of lentivirus encoding the DNA library. Cells are sorted by expression of Gene X or phenotype of interest and gRNAs within positive and negative populations are sequenced to determine enrichment of gRNAs. Enriched gRNAs bind to candidate enhancers. GWAS-SNPs within these regions can be edited for functional validation as illustrated in Key Figure, Figure 1.
