Figure 3.
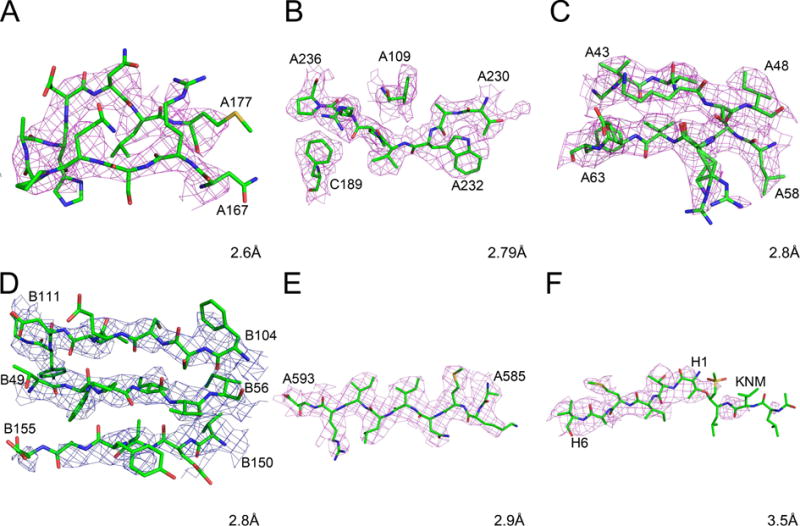
Cryo-EM maps and models for fragments of selected high-resolution protein structures. The raw maps are colored blue and the sharpened maps magenta. A) Rotavirus VP6, residues A167-A177. B) Human rhinovirus C, residues A230-A236, A109, and C189. C) Leishmania large ribosomal subunit, residues A43-A48 and A58-A63. D) Grapevine fanleaf virus complexed with a nanobody, residues B49-B54, B104-B111, and B150-B155. E) Anthrax toxin protective antigen pore, residues 585–593. F) Human 20S proteasome core, residues H1-H6 and the inhibitor KNM.
