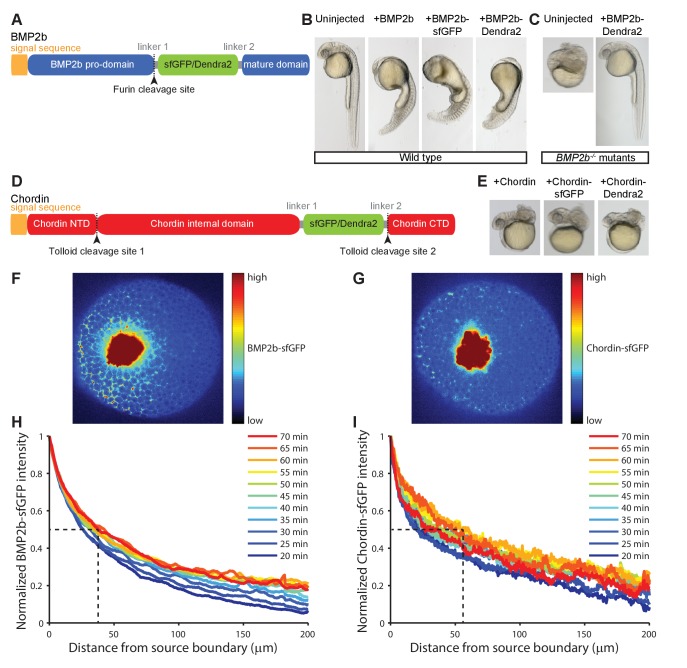
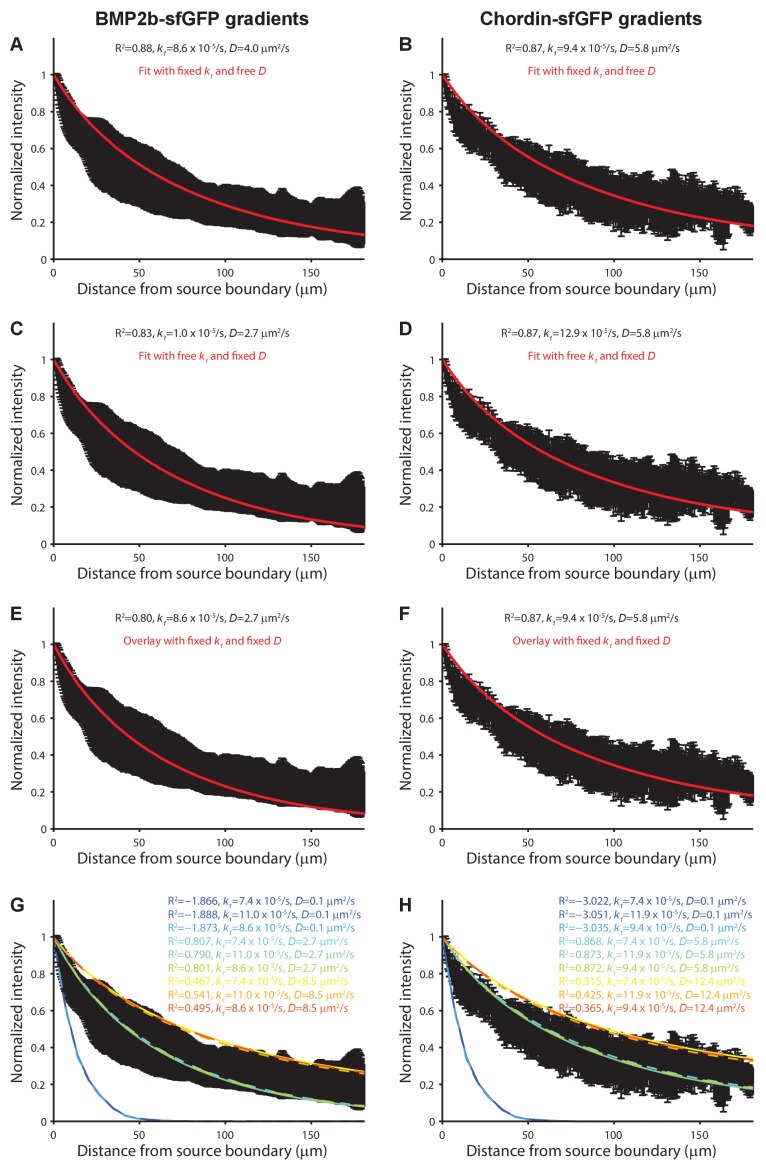
Figure 3. Gradient formation kinetics of fluorescently tagged BMP and Chordin.
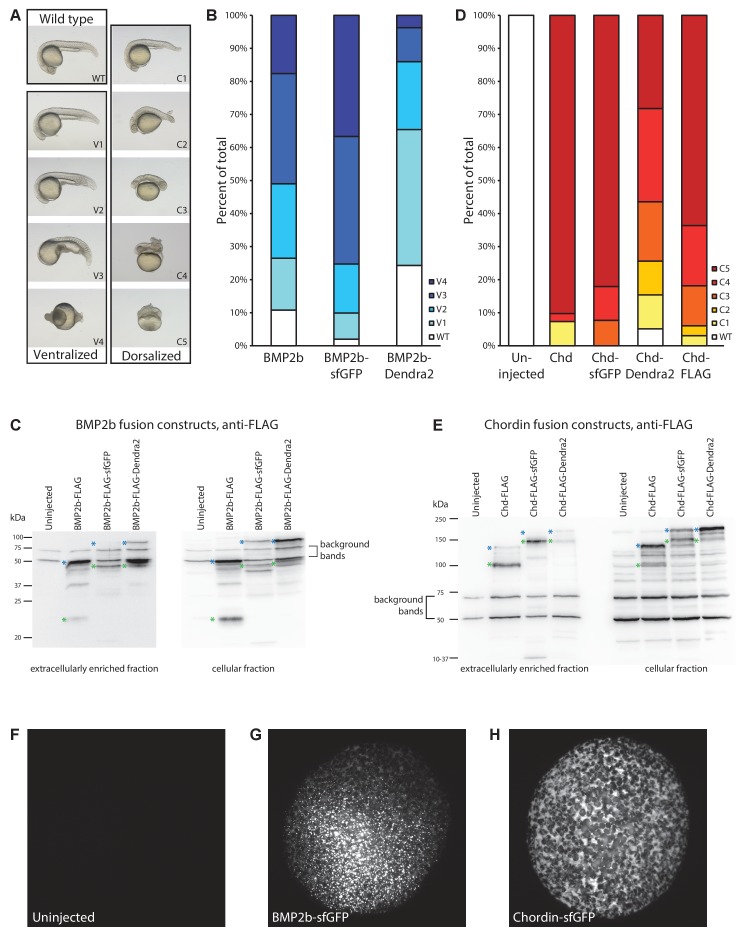
(A) Schematic of BMP2b-sfGFP and -Dendra2 fusion constructs. (B) Fluorescent BMP2b fusion constructs can induce ventralization, a BMP-overexpression phenotype (Kishimoto et al., 1997). mRNA amounts equimolar to 2 pg of BMP2b mRNA were injected at the one-cell stage, and images were taken 30 hr post-fertilization (hpf). (C) Rescue of a BMP2b mutant (swr−/−) with BMP2b-Dendra2. 2.74 pg of BMP2b-Dendra2-encoding mRNA were injected at the one-cell stage, and images were taken at 30 hpf. In a separate experiment with 1 pg of BMP2b-sfGFP-encoding mRNA, 20% (9/44) of all injected swr−/− mutants were rescued, 16% (7/44) were ventralized, and 64% (28/44) were dorsalized. (D) Schematic of Chordin-sfGFP and -Dendra2 fusion constructs. (E) Fluorescent Chordin constructs can induce dorsalization, a Chordin-overexpression phenotype. mRNA amounts equimolar to 30 pg of Chordin mRNA were injected into wild type embryos at the one-cell stage, and images were taken at 30 hpf. F + G) Light sheet microscopy images of BMP- and Chordin-sfGFP gradients forming from a local source in live zebrafish embryos. Approximately 50–75 cells expressing BMP2b-sfGFP (F) or Chordin-sfGFP (G) were transplanted into host embryos at sphere stage (see Materials and methods for details). The images show gradient formation in single optical slices approximately 20 min after transplantation. H + I) Quantification of BMP2b-sfGFP (H) and Chordin-sfGFP (I) gradient formation kinetics from a local source (BMP2b-sfGFP: n = 8; Chordin-sfGFP: n = 5). Dashed lines indicate the distance at which the protein distributions drop to 50% of their maximal concentration 60 min post-transplantation.