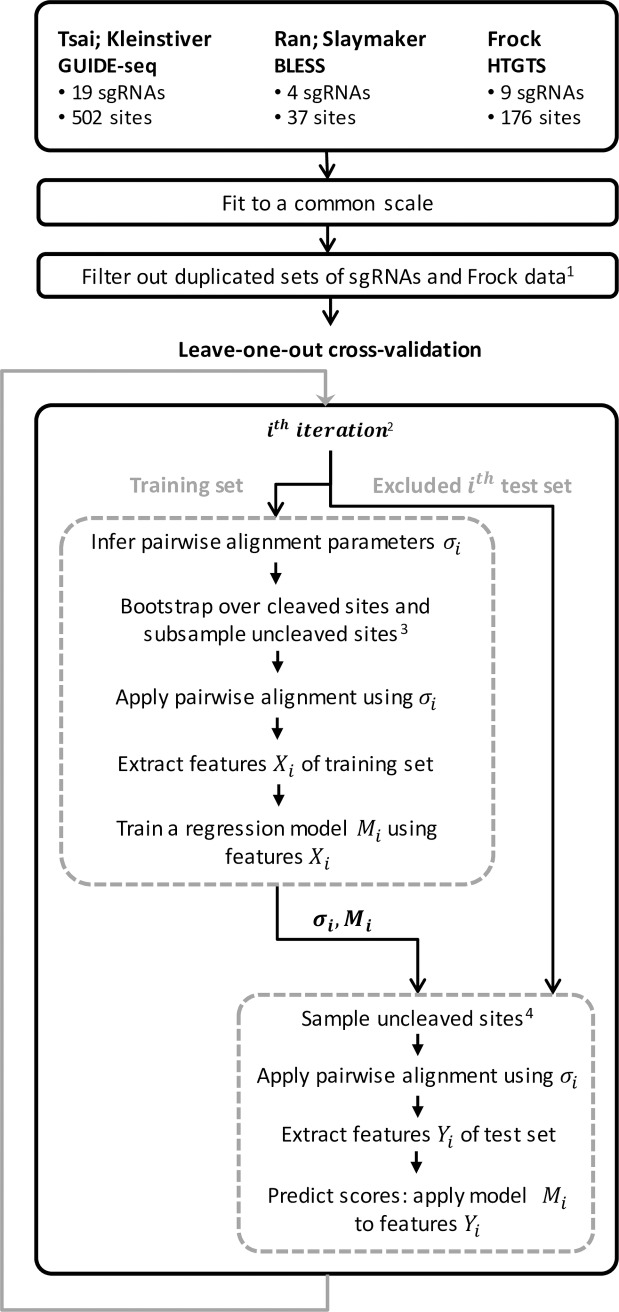
Fig 1. Schematic flow of the cross-validation procedures.
The main components of the learning pipeline for the leave-one-sgRNA-out and leave-study-out cross-validation procedures are presented. 1 This step was applied to the leave-one-sgRNA-out procedure only. 2 In each iteration, the samples of a single sgRNA (in the case of the leave-one-sgRNA-out procedure) or all samples from a single study (in the case of leave-study-out) were excluded from the training data and used as a test set. The algorithm was trained on the rest of the data. 3 Each set of cleaved samples (targets that correspond to a single sgRNA) was oversampled using bootstrapping, thus introducing a subset twice the size of the original one, and an equal-sized set of uncleaved samples was randomly chosen. 4 For each original set of cleaved samples in the test set (targets that correspond to a single sgRNA), an equal-sized set of uncleaved samples was randomly chosen.