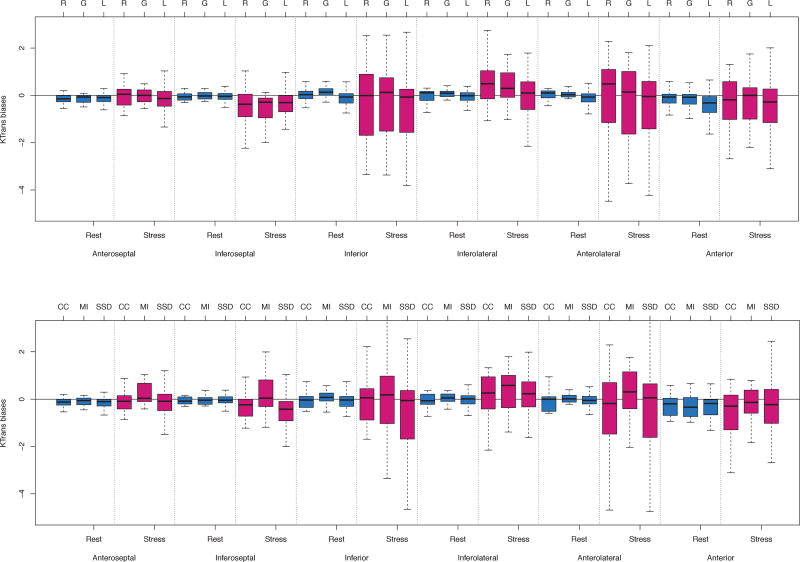
Fig. 4.
Top: The Ktrans bias difference distributions (shown by Whisker’s boxplot) between rigid (R), global affine (G) and local deformation (L) registration algorithms. Bottom: The Ktrans bias difference distributions between cross-correlation (CC), mutual information (MI) and sum-of-squared distance (SSD) registration metrics as minimisation criteria. Each figure was grouped by region and rest/stress. The thick horizontal lines inside each box indicate the median values; the boxplot height ranges from the first and third quartiles; and the hinges indicate interquartile ranges.