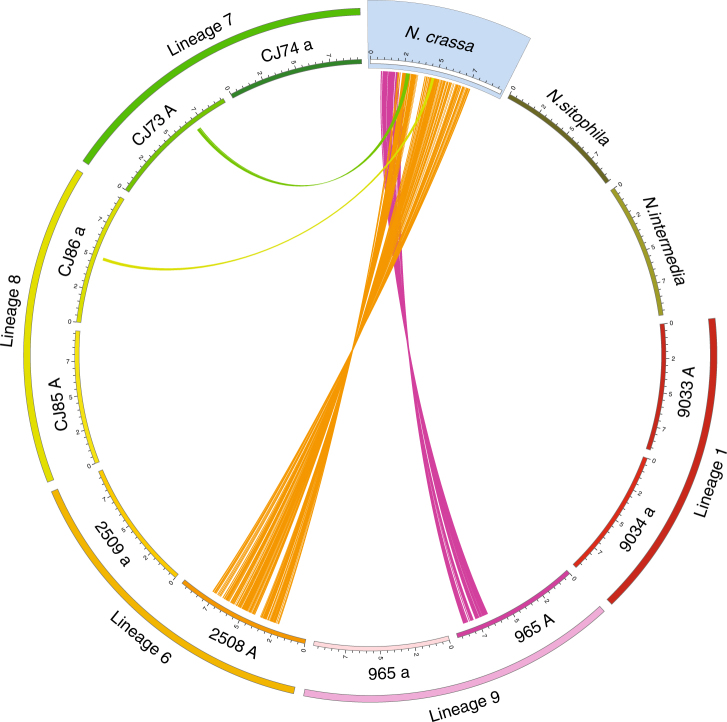
Fig. 1.
Whole chromosome pairwise alignments of the mat chromosome of N. tetrasperma against N. crassa. The mat chromosome (chromosome 1) of the 10 high-quality de novo assemblies (eight N. tetrasperma, one N. sitophila and one N. intermedia) generated for this study (Table 1), together with two mat chromosomes from Lineage 630, were aligned to the N. crassa OR74A assembly31. Chromosomes are scaled in Mbp and only inverted regions larger than 100 kbp are shown. For N. tetrasperma, genomes of each of the two mating types from a single heterokaryon were sequenced separately. Each heterokaryon represent a phylogenetically distinct lineage (Lineages 1, 6, 7, 8 and 9)27,33,34. The previously identified set of inversions in 2508 mat A (Lineage 6)30 is the biggest inverted region identified in N. tetrasperma. Strain 965 mat A (Lineage 9) carries a 1.9 Mbp inversion surrounding the mat locus, CJ86 mat a (Lineage 8) carries a 260 kbp inversion and CJ73 mat A (Lineage 7) a 320 kbp inversion. Both mating type strains of Lineage 1 completely lack large inversions on this chromosome, as do the N. sitophila and N. intermedia genomes