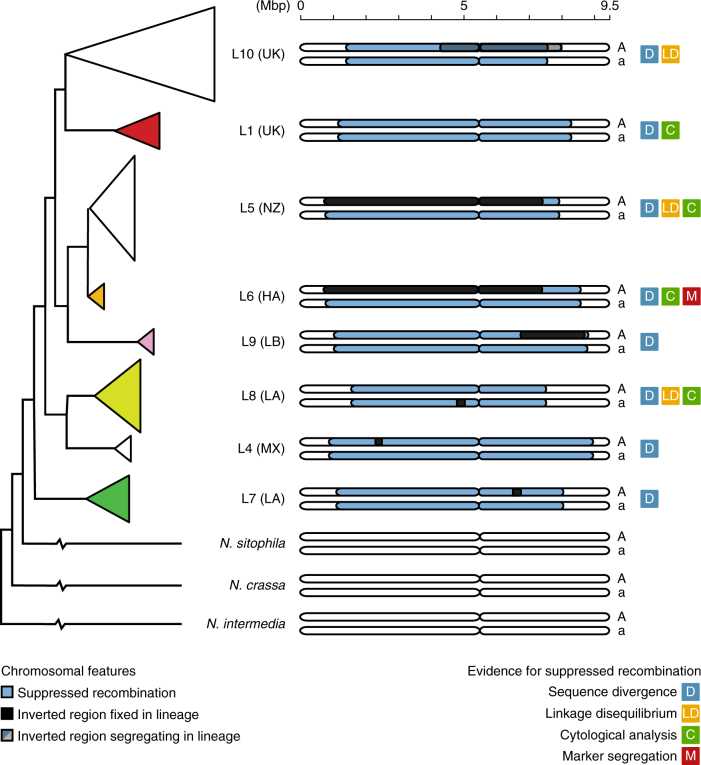
Fig. 2.
Structure of the mat chromosome across N. tetrasperma lineages. All inversions above 100 kbp on mat chromosomes identified in this study, plotted per lineage. The phylogenetic tree is adapted from Corcoran et al.27 and the colours of the filled triangles correspond to the colours used in Fig. 1. Several studies have provided different types of evidence for suppression of recombination along the mat chromosome in N. tetrasperma. Gallegos et al.26 investigated strains from Lineages 1, 5, 6 and 8 and determined both that they lack chromosomal synapsis at pachytene in most of the mat chromosome, and that no crossovers occurred between a set of genetic markers for a Lineage 6 heterokaryon. Corcoran et al.27 identified a region of elevated sequence divergence between the mating types within a heterokaryon for all N. tetrasperma lineages included in this study, as well as a region of elevated linkage disequlibirium for three of them. The combination of evidence indicates that all N. tetrasperma lineages carry a region of suppressed recombination on the mat chromosome (blue bars), but the size of this region differs between lineages27. Black bars show inverted regions that are fixed on a mating type within a lineage. All identified inverted regions are fixed in all investigated strains of the same mating type within its lineage, except a 3.6 Mbp inversion only identified in CJ01 mat A in Lineage 10. The inverted region in Lineages 5 and 6 is the only one that is shared between lineages. Neurospora sitophila, N. crassa and N. intermedia share the gene order with the majority of N. tetrasperma strains, but the mat chromosome recombines freely. This distribution of inversions shows that all identified inversions are derived and that only the suppression of recombination is shared between all analyzed strains of N. tetrasperma