Figure 3.
Structural Analysis of the OTULIN-bio-UbG76Dha-Ub Complex
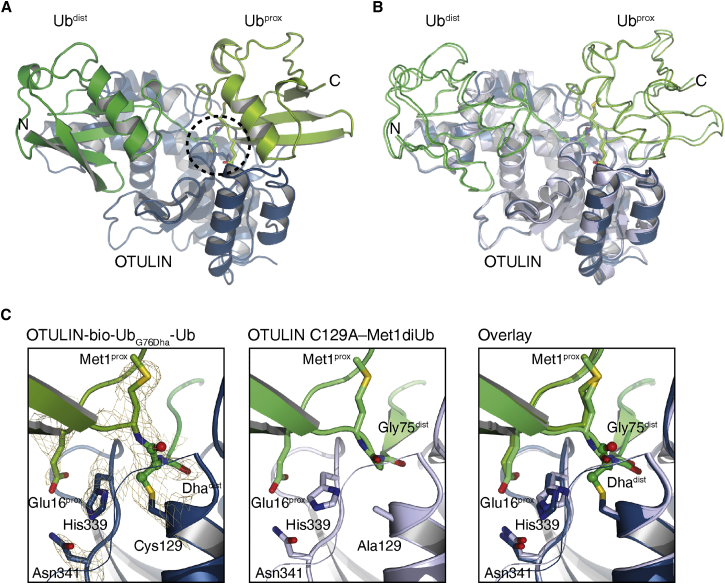
(A) Structure of the covalent OTULINcat-bio-UbG76Dha-Ub complex. OTULIN (amino acids [aa] 80–352) shown in blue with the distal and proximal ubiquitin moieties of OTULIN ABP colored in dark and light green, respectively. Circle: site of the OTULIN catalytic triad. See also Table S1.
(B) Superimposition of the OTULIN catalytic domains from the OTULIN-bio-UbG76Dha-Ub structure (dark blue) with the OTULIN C129A M1-linked diUb complex (OTULIN C129A; light blue) (PDB: 3ZNZ). The diUb moieties are shown as ribbon to highlight the near identical arrangement.
(C) Close-up views of the OTULIN-bio-UbG76Dha-Ub active site (left) and the OTULIN C129A M1-diUb complex active site (middle). Residues 118–128 are shown as ribbons. Catalytic residues are shown in stick format, as is the Glu16prox residue. The Dha group is shown additionally as spheres. A simulated annealing composite omit map, contoured at 1.2σ is shown encompassing the key catalytic residues and the Dha group. Right: superimposition of the two structures highlighting the identical arrangement of the catalytic triad.