Figure 3.
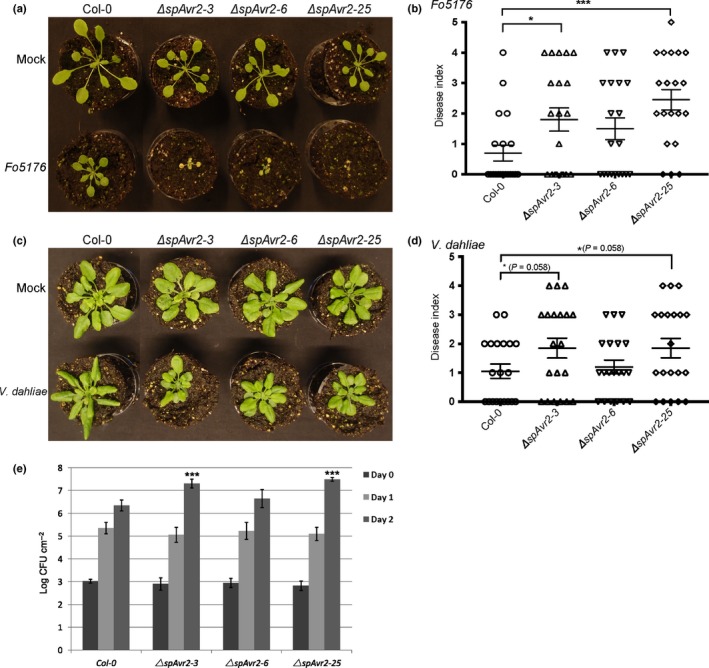
ΔspAvr2 transgenic Arabidopsis lines exhibit increased susceptibility to Fo5176, Verticillium dahliae and Pseudomonas syringae pv. tomato (Pst) DC3000. (a) Fo5176 bioassay on 14‐d‐old plants. Photographs of representative plants 10 d after mock (upper row) or Fo5176 inoculation (lower row). The result from one representative repeat from three independent biological repeats is shown. (b) Disease index 10 d post inoculation (dpi) after Fo5176 inoculation. Bars indicate ± SD of 20 replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001 using one‐way ANOVA test. (c) V. dahliae bioassay on 14‐d‐old plants, representative photographs show disease development 21 dpi with mock (upper row) or fungus (lower row). (d) Disease index 21 dpi after V. dahliae inoculation. Bars indicate ± SD of 20 replicates. The result from one representative repeat from three independent biological repeats is shown, one‐way ANOVA. (e) Pst DC3000 was infiltrated into leaves of 5‐wk‐old Arabidopsis plants. Bacterial titers were measured at 0, 1, and 2 dpi (two leaf discs per replicate, error bars indicate standard deviation of four plants/line). Three biological replicates were performed and representative data from one experiment is shown. (***, P < 0.001; t‐test).
