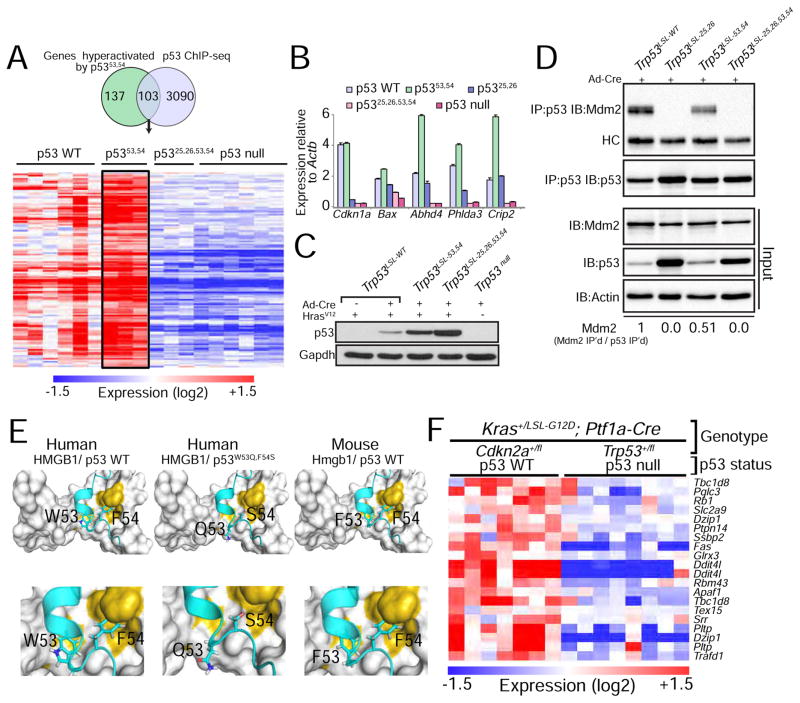
Figure 2. p53 transcriptomics and ChIP-seq analysis reveal genes hyperactivated by p5353,54.
(A) Genes activated in both HrasV12;Trp5353,54/53,54 and HrasV12;Trp53+/+ MEFs relative to HrasV12;Trp53−/− MEFs were further filtered based on fold change (HrasV12;Trp5353,54/53,54 vs. HrasV12;Trp53+/+ ≥ 1.3 fold) and on direct binding by p53, as established by p53 ChIP-seq data. The heat map shows the expression of 103 genes that satisfy these criteria in MEFs of these genotypes as well as in HrasV12;Trp5325,26,53,54/25,26,53,54 MEFs. (B) qRT-PCR analysis of p53 target gene expression in KrasG12D-expressing lung cancer cells derived from tumors in Kras+/LA2;Trp53LSL-WT/LSL-WT, Kras+/LA2;Trp53LSL-25,26/LSL-25,26, Kras+/LA2;Trp53LSL-53,54/LSL-53,54 or Kras+/LA2;Trp53LSL-25,26,53,54/LSL-25,26,53,54 mice and transduced with Ad-Cre to allow for the recombination of the lox-stop-lox cassette. Expression is relative to Actb (n=1, triplicate). Data are presented as mean ± SD. (C) Western blot analysis of p53 in HrasV12-expressing MEFs homozygous for the different p53 variants. Cells were transduced with Ad-Cre (Adenovirus-Cre) to allow for the recombination of the lox-stop-lox cassette present in the different Trp53 alleles. Gapdh is a loading control. (D) Immunoprecipitation (IP) of p53 in MEFs homozygous for different Trp53 alleles, followed by immunoblotting for Mdm2 and p53. HC denotes the immunoglobulin heavy chain. Input is 2.5% of the total amount immunoprecipitated. Actin serves as a loading control. The fraction of Mdm2 bound to each p53 variant is indicated at the bottom and is relative to wild-type p53. (E) In silico structural modeling showing how the 53,54 residues in human and mouse p53 interact with HMGB1, a representative p53 TAD2-interacting protein, and how this interaction is affected with the QS mutations. (F) Heat map showing the subset of the 103 genes identified in HrasV12-expressing MEFs whose expression is p53-dependent in PDAC, based on the comparison of PDAC cells from Kras+/LSL-G12D;Ptf1a-Cre;Trp53+/fl and Kras+/LSL-G12D;Ptf1a-Cre;Cdkn2a+/fl mice, where tumors undergo LOH for Trp53 or Cdkn2a (Collisson et al., 2011).