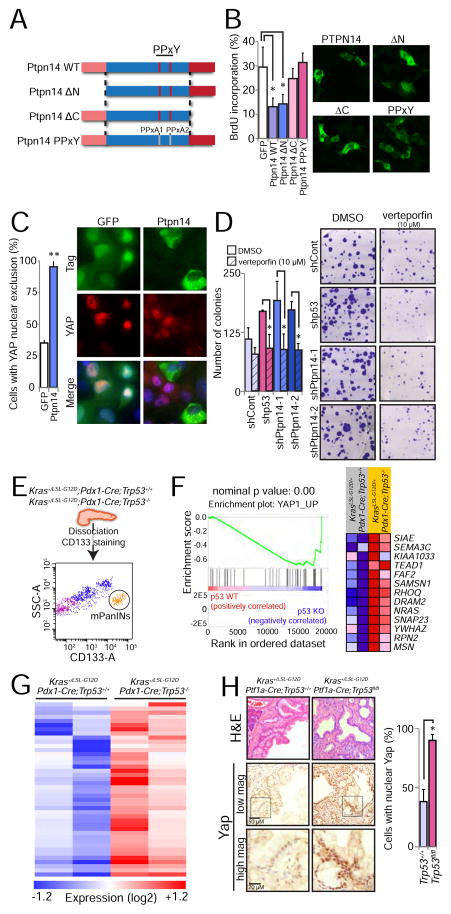
Figure 6. Ptpn14 restrains Yap activity.
(A) Schematic representation of wild-type Ptpn14 and a panel of mutants altered in key domains, including the FERM, PPxY and protein tyrosine phosphatase domains. PPxA1/PPxA2 denotes alanine mutations in both PPxY domains (Wilson et al., 2014). (B) HA-GFP or V5-tagged Ptpn14 variants were expressed in KPC mouse PDAC cells and BrdU-positivity of cells expressing each antigen (detected by GFP, HA or V5 immunofluorescence) was assessed. (Left) The average BrdU incorporation from 3 experiments ± SD, with 100 cells counted per experiment, is shown. (Right) Representative immunofluorescence images showing the localization pattern for each Ptpn14 variant. (C) Effects of HA-Ptpn14 on YAP localization in PANC-1 human PDAC cells, compared to HA-GFP. (Left) Graph showing the average percentage of cells with YAP nuclear exclusion from 3 experiments ± SD. (Right) Representative images with arrows pointing to HA-GFP-positive cells with nuclear YAP and arrowheads pointing to Ptpn14-expressing cells without nuclear Yap. (D) Colony forming ability of KIC cells treated with the Yap inhibitor verteporfin (10 μM) after introduction of control, p53, Ptpn14-1 or Ptpn14-2 shRNAs. (Left) Low plating experiment quantification, showing the average colony number ± SD of 3 experiments. (Right) Representative images show the crystal violet-stained colonies 2 weeks after seeding. (E) Sorting scheme to isolate PanIN cells from Kras+/G12D;Pdx1-Cre;Trp53+/+ and Kras+/G12D;Pdx1-Cre;Trp53−/− mice, based on CD133 positivity. Side-scatter area (SSC-A) and CD133 area (CD133-A) are used to define the PanIN population. (F) Yap GSEA signature (YAP1_UP) found enriched in Kras+/G12D;Pdx1-Cre;Trp53−/− cells relative to Kras+/G12D;Pdx1-Cre;Trp53+/+ cells. Nominal p value is indicated. The heat map represents the expression of the genes that contributed to the enrichment score. (G) Heat map showing the expression of an expanded list of 114 Yap-activated target genes based on 3 different signatures in Kras+/G12D;Pdx1-Cre;Trp53+/+ and Kras+/G12D;Pdx1-Cre;Trp53−/− PanIN cells. (H) (Left) Representative histological images of PanIN lesions from Kras+/G12D;Ptf1a-Cre;Trp53+/+ (n=4) and Kras+/G12D;Ptf1a-Cre;Trp53fl/fl (n=3) mice. The same field was analyzed by H&E and Yap staining, at 2 different magnifications, as indicated by the scale bars. Box indicates region of magnification. (Right) Average percentage of cells in PanINs with nuclear Yap ± SD. At least 300 nuclei were analyzed per mouse. * p ≤ 0.05; ** p ≤ 0.001, two-tailed unpaired Student’s t-test.