FIGURE 3.
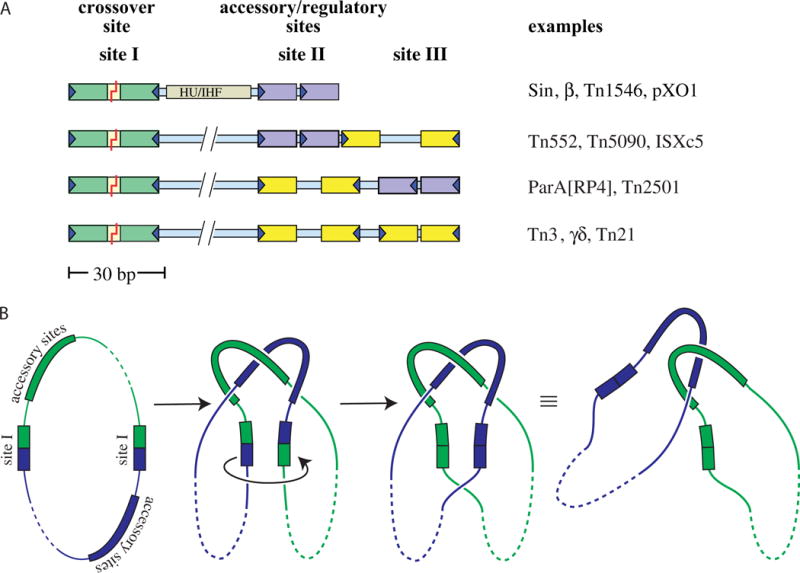
Res sites and the topology of the synaptosome. (A) Examples of serine resolvase res sites. Specific recognition sequences (~12 bp each) for individual resolvase subunits are shown as colored boxes: green for the crossover site (always an inverted repeat), purple for accessory sites that form direct repeats, and yellow for accessory sites that form inverted repeats. The recombinase dimers bound to the accessory sites are catalytically inactive, and these sites always differ from the crossover site in the length of their central spacers and/or the relative orientation of their half-sites. Sin and related resolvases require a DNA bending protein as well as additional recombinase subunits. The coding sequence for the resolvase protein is usually adjacent to its cognate res site. Figure adapted from reference (60) with permission. (B) Resolvase synaptosomes trap 3 supercoiling nodes. The resolvase and accessory proteins (if any) bound to each of the cognate res sites form a complex (the “synaptosome”) that traps 3 dsDNA-over-dsDNA crossings and juxtaposes the two site Is. Synaptosome formation activates the site I-bound resolvase subunits, which then introduce double-strand breaks. A 180° rotation of the bottom two subunits (as drawn) realigns the broken ends, which are then re-ligated. In a negatively supercoiled substrate a right-handed rotation is favored because it introduces a + supercoiling node (ΔLk = +1) that cancels one of the pre-existing (−) nodes trapped in the synaptosome and because it allows rewinding of each duplex by a half turn (ΔTw for each = +½). The remaining two crossings trapped by the synaptosome are no longer intramolecular and instead catenate the two daughter circles (which can be separated by a host type II topoisomerase). doi:10.1128/microbiolspec.MDNA3-0045-2014.f3 31)). (B) Activated mutant γδ resolvase with crossover site DNA in the covalent protein–DNA intermediate state. Note that each catalytic domain has undergone major conformational changes in the transition from dimer to tetramer (PDBid 2gm4; (48, 49)). (C) The same structure as in B, rotated by ~90° about a horizontal axis. (D) Activated Sin resolvase tetramer catalytic domain tetramer. Sulfate ions that mark the binding pockets for the scissile phosphate are shown as sticks (PDBid 3pkz) (50). doi:10.1128/microbiolspec.MDNA3-0045-2014.f4
