FIGURE 7.
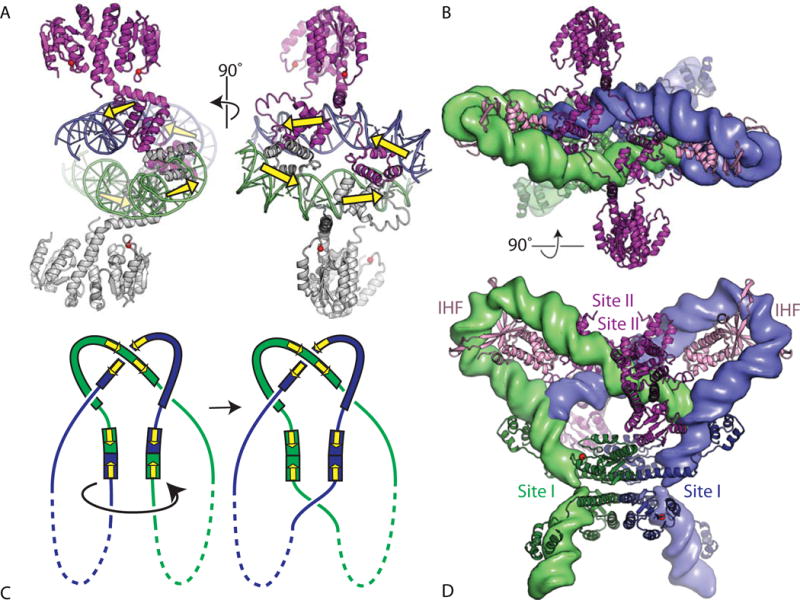
Modeling the Sin synaptosome. (A) Two orthogonal views of the structure of a tetramer of WT Sin resolvase bound to its cognate site II. In the left panel, the catalytic domains of the lower dimer are oriented in a similar way to those of wild-type γδ resolvase in Figure 4(A). The dimer–dimer contacts are mediated by the DNA-binding domains (PDBid 2r0q (64)). Yellow arrows show the orientations of the individual monomer-binding sites in the DNA. (B) Model for the synaptosome, created by rigid-body docking together of a symmetrized version of an activated γδ resolvase tetramer–DNA structure (blue and green proteins), two copies of an IHF–DNA complex structure (pink proteins), and the Sin–site II structure (purple proteins). The DNAs are shown as smoothed green and blue surfaces. The view is the same as for the right hand panel of (A). PDBids 1zr4, 1ihf, and 2r0q were used (48, 64, 84). (C) Cartoon similar to that in Figure 3B showing the expected synaptosome topology. Yellow arrows mark the orientations of the half-sites within each dimer-binding site. (D) Second view of the synaptosome model, rotated 90° about a horizontal axis. doi:10.1128/microbiolspec.MDNA3-0045-2014.f7 50)). A sulfate ion marks the scissile-phosphate binding pocket, and side chains important for catalysis are shown as sticks. Those residues whose mutation had the most deleterious effect on the rate of DNA cleavage by Tn3 resolvase are shown in magenta, shading to white for those whose mutation had more moderate effects (74). (B) Stereo view of one subunit from a site II-bound wild-type Sin dimer (PDBid 2r0q (64)). The same side chains are shown, similarly shaded from a dark color to white. doi:10.1128/microbiolspec.MDNA3-0045-2014.f9
