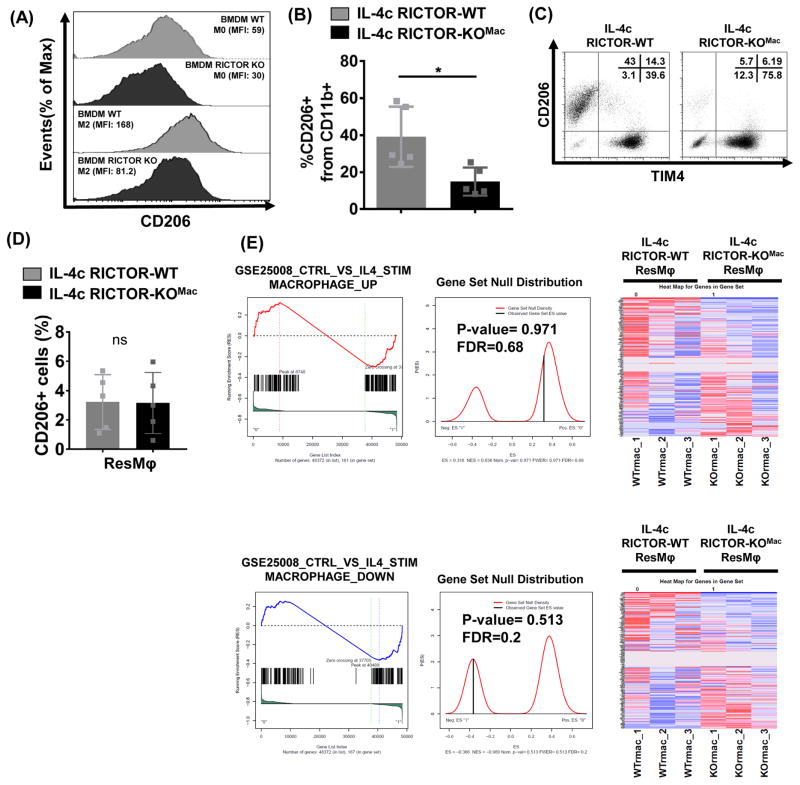
Figure 3. mTORC2 activity differentially regulate IL-4c driven alternatively activated macrophages and tissue-resident macrophages.
(A) BMDM from RICTOR-WT and RICTOR-KOMac mice and were cultured with 10ng/mL IL-4 for 18hrs. CD206 was measured by flow cytometry. (B–D) Peritoneal exudate cells were collected from IL-4c treated RICTOR-WT and RICTOR-KOMac mice. (B) Percentages of CD206+ cells from CD11b+ population (N=5/group). (C) Representative flow analysis of TIM4 and CD206 expression on CD11b+ population. Percentages are shown in upper right corner. (D) Flow analysis of CD206+ cells among ResMφ (CD11b+TIM4+F4/80hi). (E) Gene-set enrichment analysis from RNA sequencing on RICTOR-WT and RICTOR-KOMac peritoneal ResMφ (CD11b+TIM4+ F4/80hi) (2–3 mice pooled N=3). Presented as enrichment for selected pathways among genes previously upregulated (top) or downregulated (bottom) from BMDM control vs. IL-4 gene set (GSE25088). Data are representative of at least three independent experiments. *P < 0.05, Mann-Whitney t tests.