Figure 2. Genomic and protein structure of GLE1 and spectrum of pathogenic mutations in GLE1.
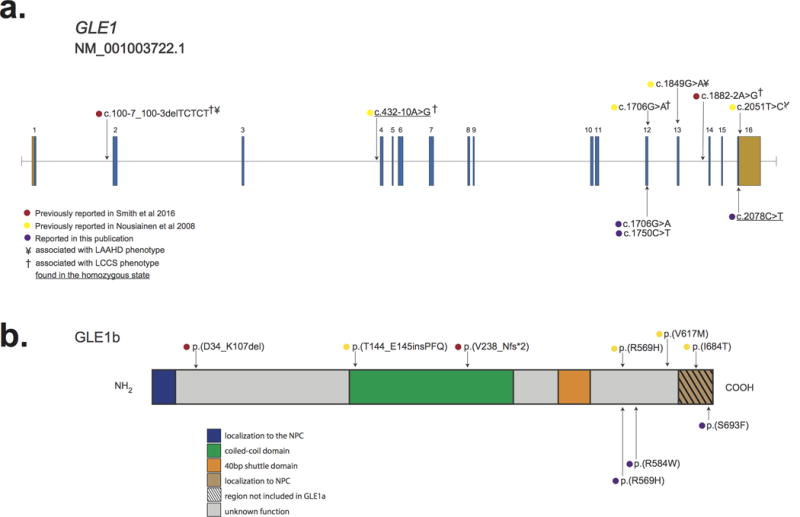
a.) GLE1 is composed of 16 exons, all which are protein-coding (blue) and two of which also contain non-coding sequence (orange). Lines with attached dots indicate the approximate locations of eleven different recessive variants that cause LCCS1 and LAAHD. The color of each dot reflects the report of each mutation. b.) Protein topology of GLE is comprised of six domains. Most mutations lie in the C-terminal portion of GLE1. No mutations in the N-terminal Nup155 binding domain or in the nucleocytoplasmic shuttling domain have been reported. The approximate positions of variants that cause LCCS1 and LAAHD are indicated by colored dots. Only p.(V238_Nfs*2) has been shown to result in complete lack of GLE1; all other variants have been shown or are expected to be expressed.
