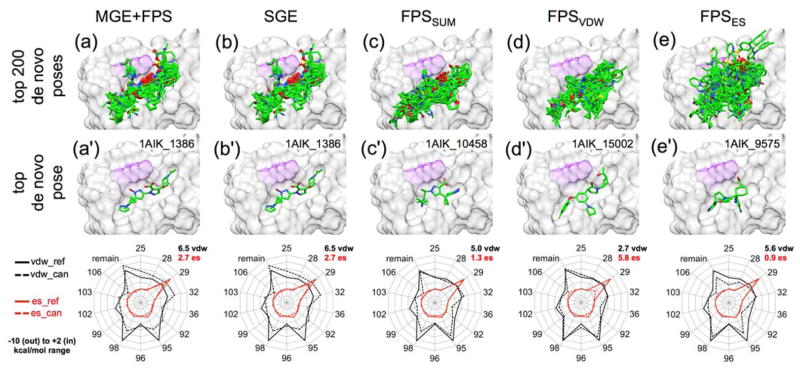
Figure 14.
Examples of molecules constructed in the HIV gp41 hydrophobic pocket through de novo design using the MGE+FPS scoring function and rescored using five different functions: MGE+FPS, SGE, FPSSUM, FPSVDW, and FPSES. Top panels show the top 200 scoring molecules for each ranking metric. Middle panels show the best scoring molecule for each ranking metric. Bottom panel radar plots show reference footprints (bold lines) for comparison with candidate footprints (dashed line) from the best scoring molecules. Hydrophobic pocket of gp41 in gray surface with key lysine residue highlighted in purple. Footprint similarity scores (Euclidian distance) for van der Waals (VDW) overlap and electrostatic overlap (ES) are shown in black and red font respectively.