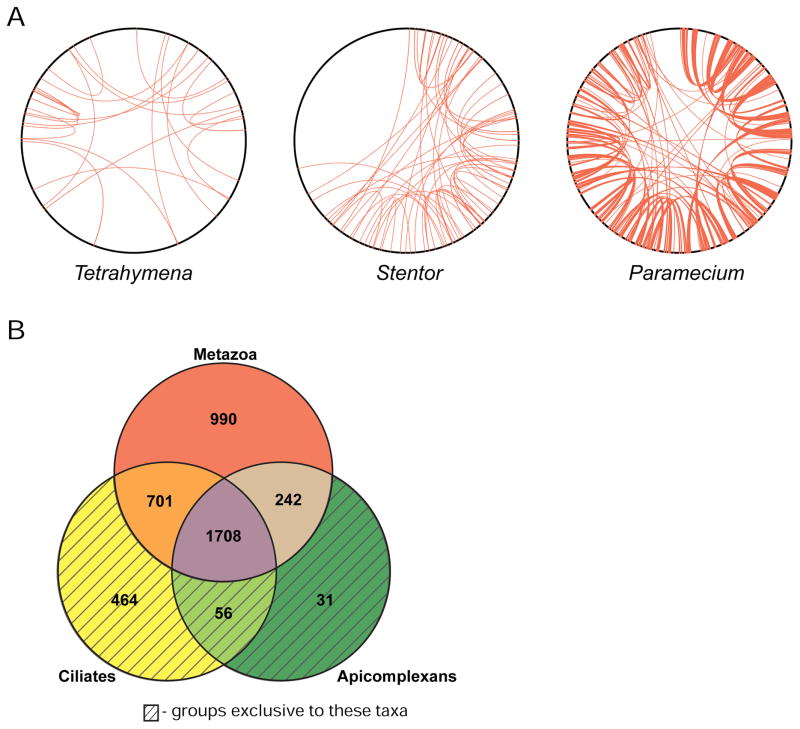
Figure 2. Stentor gene duplications and orthology groups.
(A) Genome duplication events in the genomes of Stentor coeruleus, Paramecium tetraurelia, and Tetrahymena thermophila. To generate coordinates on the perimeter of each circle, contigs/scaffolds were arranged from longest to shortest and then continuously numbered from 1 to the end of the assemblies. Red lines connect paralogous windows (see methods) between two scaffolds and indicate putative genome duplication events. (B) Venn diagram showing numbers of orthologous gene groups in Stentor that are also found in other ciliates, apicomplexans, or metazoans. Shaded regions indicate gene groups that are exclusive to those taxa; for example, the ciliate only region of the diagram represents gene groups that aren’t found in any other taxa. An additional 555 curated groups are shared with other organisms but not pictured in this diagram. See also Figure S2 and Table S2.