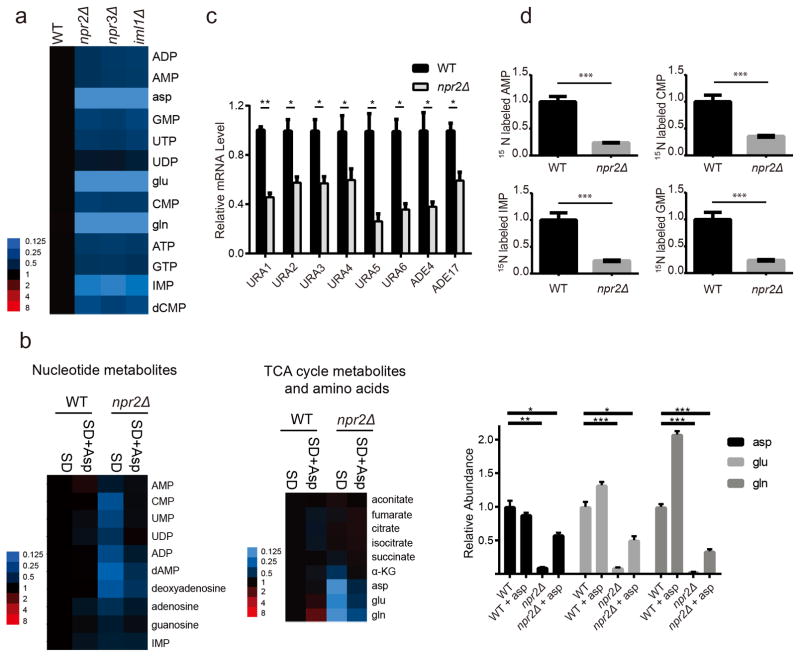
Figure 4. npr2Δ cells produce less nucleotide metabolites.
(a) Heat map depicting abundance of nucleotide metabolites in WT, npr2Δ, npr3Δ and iml1Δ cells. Cells were grown in YPD and then transferred into SD for 6 h prior to extraction. Intracellular nucleotide metabolites levels were measured with targeted LC-MS/MS methods. Data were collected from 3 independent experiments.
(b) Heat map of relative abundance of nucleotide metabolites (left) or TCA cycle metabolites, glutamine, glutamate and aspartate (middle) following aspartate supplementation, normalized against WT. Cells were switched into SD for 6 h as in (a), but with or without addition of 2 mM aspartate. Metabolites were extracted and analyzed by LC-MS/MS. Aspartate, glutamate and glutamine levels are also shown in bar graph format (right). Data were mean ± s.d. from 3 independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed Student’s t-test.
(c) mRNA levels of pyrimidine synthesis pathway enzymes (URA1, URA2, URA3, URA4, URA5, URA6) and purine synthesis pathway (ADE4, ADE17) from WT and npr2Δ cells collected as shown in (a). Data were mean ± s.d. from 2 independent experiments with duplicates for each genotype. *p < 0.05, **p < 0.01, by two-tailed Student’s t-test.
(d) Relative abundance of labeled nucleotide metabolites from 15N-ammonium tracing experiments. Cells were grown in YPD and then transferred into SD for 3 h, and then switched into SD-N plus 5 g/L ammonium-15N2 sulfate for 1 h. Data were mean ± s.d. from 3 independent experiments. ***p<0.001, by two-tailed Student’s t-test.