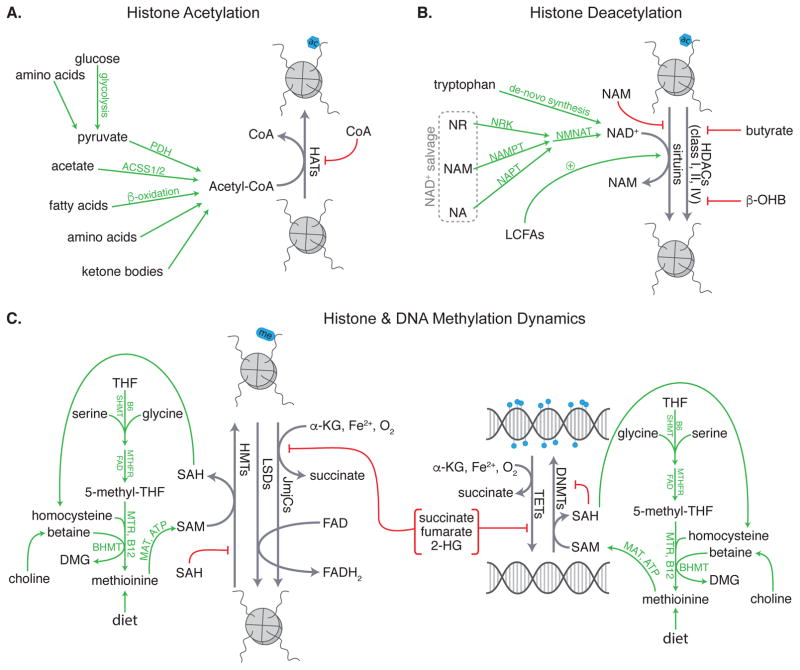
Figure 1.
Endogenous metabolites regulate (A) histone acetylation, (B) histone deacetylation, and (C) histone and DNA methylation and demethylation. PDH = pyruvate dehydrogenase, ACSS1/2 = acetyl-CoA synthetase 1/2, HAT = histone acetyltransferase, NR = nicotinamide riboside, NA = nicotinic acid, NAM = nicotinamide, NMNAT = nicotinamide/nicotinic acid mononucleotide adenylyltransferase, LCFAs = long-chain fatty acids, NAD+ = nicotinamide adenine dinucleotide, HDACs = histone deacetylases, β-OHB = β-hydroxybutyrate, SHMT = serine hydroxymethyltransferase, MTHFR = methylenetetrahydrofolate reductase, 5-methyl-THF = 5-methyl- tetrahydrofolate, THF = tetrahydrofolate, MTR = methionine synthase, DMG = dimethylglycine, MAT = S-adenosylmethionine synthase, SAH = S-adenosyl homocysteine, SAM = S-adenosylmethionine, HMTs = histone methyltransferases, LSDs = lysine specific demethylases, JmjCs = Jumonji C family histone demethylases, α-KG = α-ketoglutarate, TETs = Ten Eleven Translocation methylcytosine dioxygenases, DNMTs = DNA methyltransferases